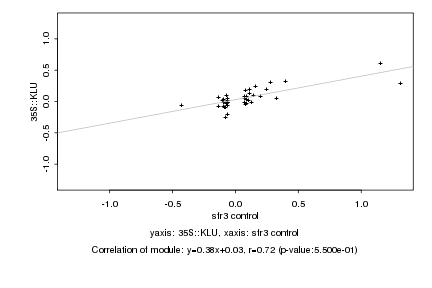
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
sfr3 control |
Log2 signal ratio
35S::KLU |
| 1 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
1.309 |
0.290 |
| 2 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
1.152 |
0.611 |
| 3 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.391 |
0.325 |
| 4 |
254700_at |
AT4G18000
|
unknown |
0.319 |
0.059 |
| 5 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.278 |
0.312 |
| 6 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.241 |
0.201 |
| 7 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
0.194 |
0.086 |
| 8 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.156 |
0.250 |
| 9 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.139 |
0.110 |
| 10 |
249055_at |
AT5G44460
|
CML43, calmodulin like 43 |
0.124 |
-0.015 |
| 11 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
0.109 |
0.192 |
| 12 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.105 |
0.140 |
| 13 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.101 |
0.031 |
| 14 |
266000_at |
AT2G24180
|
CYP71B6, cytochrome p450 71b6 |
0.086 |
0.095 |
| 15 |
259899_at |
AT1G71210
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.084 |
-0.017 |
| 16 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
0.082 |
0.045 |
| 17 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
0.073 |
0.186 |
| 18 |
257120_at |
AT3G20200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.073 |
-0.038 |
| 19 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.069 |
0.064 |
| 20 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.068 |
0.084 |
| 21 |
265718_at |
AT2G03340
|
WRKY3, WRKY DNA-binding protein 3 |
0.067 |
-0.001 |
| 22 |
261639_at |
AT1G50010
|
TUA2, tubulin alpha-2 chain |
-0.433 |
-0.055 |
| 23 |
248423_at |
AT5G51670
|
unknown |
-0.140 |
-0.064 |
| 24 |
259596_at |
AT1G28130
|
GH3.17 |
-0.137 |
0.079 |
| 25 |
246079_at |
AT5G20450
|
unknown |
-0.104 |
0.023 |
| 26 |
260809_at |
AT1G43730
|
[RNA-directed DNA polymerase (reverse transcriptase)-related family protein] |
-0.100 |
-0.004 |
| 27 |
257102_at |
AT3G25050
|
XTH3, xyloglucan endotransglucosylase/hydrolase 3 |
-0.099 |
0.035 |
| 28 |
266163_at |
AT2G28130
|
unknown |
-0.095 |
-0.076 |
| 29 |
259958_at |
AT1G53730
|
SRF6, STRUBBELIG-receptor family 6 |
-0.087 |
-0.082 |
| 30 |
257202_at |
AT3G23750
|
BARK1, BAK1-Associating Receptor-Like Kinase 1 |
-0.083 |
-0.081 |
| 31 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
-0.081 |
-0.240 |
| 32 |
261714_at |
AT1G18510
|
TET16, tetraspanin 16 |
-0.078 |
-0.023 |
| 33 |
265951_at |
AT2G18500
|
ATOFP7, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 7, OFP7, ovate family protein 7 |
-0.072 |
-0.018 |
| 34 |
255538_at |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-0.072 |
0.105 |
| 35 |
256646_at |
AT3G13590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.071 |
-0.014 |
| 36 |
254511_at |
AT4G20220
|
[Reverse transcriptase (RNA-dependent DNA polymerase)] |
-0.071 |
0.020 |
| 37 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.071 |
-0.195 |
| 38 |
247219_at |
AT5G64920
|
CIP8, COP1-interacting protein 8 |
-0.070 |
-0.014 |
| 39 |
256054_at |
AT1G07120
|
unknown |
-0.070 |
0.049 |
| 40 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.070 |
-0.063 |
| 41 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.070 |
-0.004 |