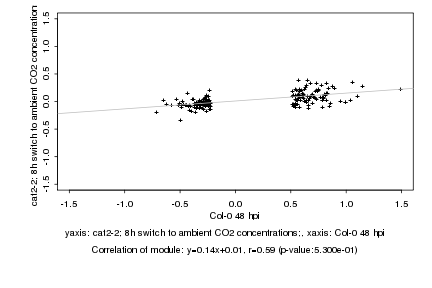
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 48 hpi |
Log2 signal ratio
cat2-2; 8h switch to ambient CO2 concentrations; |
| 1 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
1.487 |
0.224 |
| 2 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.139 |
0.273 |
| 3 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
1.094 |
0.107 |
| 4 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.055 |
0.353 |
| 5 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
1.031 |
0.033 |
| 6 |
267345_at |
AT2G44240
|
unknown |
0.991 |
-0.001 |
| 7 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.991 |
-0.010 |
| 8 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.943 |
0.017 |
| 9 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.890 |
0.238 |
| 10 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.869 |
0.278 |
| 11 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.857 |
-0.030 |
| 12 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.848 |
-0.089 |
| 13 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.833 |
0.246 |
| 14 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.823 |
0.161 |
| 15 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.820 |
0.149 |
| 16 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.819 |
0.136 |
| 17 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.816 |
0.330 |
| 18 |
258362_at |
AT3G14280
|
unknown |
0.816 |
0.024 |
| 19 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.810 |
0.118 |
| 20 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.798 |
0.137 |
| 21 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.795 |
0.088 |
| 22 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.787 |
0.058 |
| 23 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.786 |
0.080 |
| 24 |
256569_at |
AT3G19550
|
unknown |
0.782 |
-0.108 |
| 25 |
258209_at |
AT3G14060
|
unknown |
0.781 |
0.027 |
| 26 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.779 |
0.028 |
| 27 |
256937_at |
AT3G22620
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.775 |
0.292 |
| 28 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.762 |
0.081 |
| 29 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.749 |
0.220 |
| 30 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.747 |
0.207 |
| 31 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.732 |
0.191 |
| 32 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.729 |
0.343 |
| 33 |
258203_at |
AT3G13950
|
unknown |
0.727 |
0.207 |
| 34 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.726 |
0.047 |
| 35 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.722 |
0.056 |
| 36 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.717 |
0.188 |
| 37 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.710 |
0.081 |
| 38 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.704 |
0.090 |
| 39 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.693 |
0.142 |
| 40 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.687 |
-0.026 |
| 41 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
0.683 |
0.114 |
| 42 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.677 |
0.328 |
| 43 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.671 |
0.103 |
| 44 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
0.660 |
0.055 |
| 45 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.657 |
0.033 |
| 46 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.653 |
-0.114 |
| 47 |
257154_at |
AT3G27210
|
unknown |
0.652 |
-0.066 |
| 48 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.643 |
0.384 |
| 49 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.642 |
0.106 |
| 50 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.641 |
0.298 |
| 51 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.637 |
-0.017 |
| 52 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.636 |
0.024 |
| 53 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.628 |
0.260 |
| 54 |
247882_at |
AT5G57785
|
unknown |
0.620 |
0.007 |
| 55 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.617 |
0.234 |
| 56 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.614 |
0.144 |
| 57 |
249454_at |
AT5G39520
|
unknown |
0.613 |
0.114 |
| 58 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.611 |
0.058 |
| 59 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.605 |
0.119 |
| 60 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.601 |
0.161 |
| 61 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.597 |
0.088 |
| 62 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.588 |
0.202 |
| 63 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.581 |
0.025 |
| 64 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.578 |
0.194 |
| 65 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.578 |
0.235 |
| 66 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.573 |
0.118 |
| 67 |
263772_at |
AT2G06255
|
ELF4-L3, ELF4-like 3 |
0.571 |
-0.098 |
| 68 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.570 |
0.081 |
| 69 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.568 |
0.390 |
| 70 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.567 |
0.126 |
| 71 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.566 |
0.142 |
| 72 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.561 |
0.059 |
| 73 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.561 |
0.124 |
| 74 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.560 |
0.116 |
| 75 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.560 |
-0.037 |
| 76 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.557 |
0.034 |
| 77 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.549 |
0.200 |
| 78 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.544 |
0.019 |
| 79 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
0.542 |
0.046 |
| 80 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.541 |
-0.069 |
| 81 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.541 |
0.105 |
| 82 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.540 |
0.236 |
| 83 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.535 |
-0.095 |
| 84 |
251789_at |
AT3G55450
|
PBL1, PBS1-like 1 |
0.535 |
-0.021 |
| 85 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.535 |
0.126 |
| 86 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.524 |
0.124 |
| 87 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.519 |
0.120 |
| 88 |
257697_at |
AT3G12700
|
NANA, NANA |
0.517 |
-0.045 |
| 89 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.516 |
-0.075 |
| 90 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.510 |
0.185 |
| 91 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.509 |
0.105 |
| 92 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.508 |
-0.040 |
| 93 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.719 |
-0.186 |
| 94 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.650 |
0.026 |
| 95 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
-0.626 |
-0.037 |
| 96 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-0.579 |
-0.058 |
| 97 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.533 |
0.037 |
| 98 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.523 |
-0.065 |
| 99 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.510 |
-0.031 |
| 100 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.505 |
-0.339 |
| 101 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.492 |
-0.058 |
| 102 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.490 |
-0.107 |
| 103 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.486 |
0.009 |
| 104 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.478 |
-0.038 |
| 105 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.458 |
-0.055 |
| 106 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.448 |
-0.066 |
| 107 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
-0.442 |
-0.066 |
| 108 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.439 |
0.149 |
| 109 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.430 |
-0.081 |
| 110 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.421 |
-0.062 |
| 111 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.415 |
-0.161 |
| 112 |
247878_at |
AT5G57760
|
unknown |
-0.410 |
-0.083 |
| 113 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
-0.404 |
-0.176 |
| 114 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
-0.388 |
0.037 |
| 115 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.382 |
0.045 |
| 116 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.378 |
-0.096 |
| 117 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.377 |
-0.071 |
| 118 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.373 |
-0.020 |
| 119 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
-0.362 |
-0.191 |
| 120 |
247474_at |
AT5G62280
|
unknown |
-0.360 |
-0.014 |
| 121 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.357 |
-0.033 |
| 122 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.355 |
-0.046 |
| 123 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.354 |
-0.115 |
| 124 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.353 |
-0.103 |
| 125 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
-0.350 |
-0.069 |
| 126 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.341 |
-0.025 |
| 127 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.336 |
-0.009 |
| 128 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.333 |
-0.064 |
| 129 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.332 |
-0.115 |
| 130 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.330 |
0.010 |
| 131 |
251059_at |
AT5G01810
|
ATPK10, PROTEIN KINASE 10, CIPK15, CBL-interacting protein kinase 15, PKS3, SIP2, SOS3-INTERACTING PROTEIN 2, SNRK3.1, SNF1-RELATED PROTEIN KINASE 3.1 |
-0.329 |
0.033 |
| 132 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.321 |
-0.049 |
| 133 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.319 |
-0.064 |
| 134 |
246885_at |
AT5G26230
|
MAKR1, membrane-associated kinase regulator 1 |
-0.318 |
-0.091 |
| 135 |
245190_at |
AT1G67690
|
[Zincin-like metalloproteases family protein] |
-0.309 |
0.016 |
| 136 |
253533_at |
AT4G31590
|
ATCSLC05, CELLULOSE-SYNTHASE LIKE C5, ATCSLC5, Cellulose-synthase-like C5, CSLC05, CELLULOSE-SYNTHASE LIKE C5, CSLC5, Cellulose-synthase-like C5 |
-0.302 |
-0.006 |
| 137 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.302 |
-0.036 |
| 138 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.301 |
-0.109 |
| 139 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
-0.297 |
-0.131 |
| 140 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.297 |
0.010 |
| 141 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.293 |
-0.025 |
| 142 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
-0.292 |
0.049 |
| 143 |
249541_at |
AT5G38130
|
[HXXXD-type acyl-transferase family protein] |
-0.291 |
-0.028 |
| 144 |
257959_at |
AT3G25560
|
AtNIK2, NIK2, NSP-interacting kinase 2 |
-0.288 |
0.055 |
| 145 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.286 |
-0.051 |
| 146 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.286 |
-0.035 |
| 147 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.285 |
-0.026 |
| 148 |
266295_at |
AT2G29550
|
TUB7, tubulin beta-7 chain |
-0.285 |
-0.002 |
| 149 |
261605_at |
AT1G49580
|
[Calcium-dependent protein kinase (CDPK) family protein] |
-0.283 |
-0.071 |
| 150 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.279 |
-0.090 |
| 151 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
-0.278 |
-0.088 |
| 152 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.278 |
-0.010 |
| 153 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.278 |
-0.062 |
| 154 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.275 |
-0.049 |
| 155 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.272 |
0.090 |
| 156 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.271 |
-0.076 |
| 157 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.271 |
-0.057 |
| 158 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.269 |
0.122 |
| 159 |
248597_at |
AT5G49160
|
DDM2, DECREASED DNA METHYLATION 2, DMT01, DNA METHYLTRANSFERASE 01, DMT1, DNA METHYLTRANSFERASE 1, MET1, methyltransferase 1, MET2, METHYLTRANSFERASE 2, METI, METHYLTRANSFERASE I |
-0.268 |
-0.052 |
| 160 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.268 |
0.049 |
| 161 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.267 |
-0.170 |
| 162 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.266 |
0.027 |
| 163 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.264 |
-0.002 |
| 164 |
246231_at |
AT4G37080
|
unknown |
-0.257 |
0.018 |
| 165 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
-0.256 |
-0.000 |
| 166 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.255 |
0.010 |
| 167 |
245171_at |
AT2G47560
|
[RING/U-box superfamily protein] |
-0.252 |
-0.005 |
| 168 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.252 |
-0.089 |
| 169 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.250 |
0.022 |
| 170 |
266439_s_at |
AT2G43200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.249 |
-0.051 |
| 171 |
266921_at |
AT2G45970
|
CYP86A8, cytochrome P450, family 86, subfamily A, polypeptide 8, LCR, LACERATA |
-0.248 |
-0.057 |
| 172 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.246 |
0.020 |
| 173 |
258884_at |
AT3G10050
|
OMR1, L-O-methylthreonine resistant 1 |
-0.245 |
-0.049 |
| 174 |
262026_at |
AT1G35670
|
ATCDPK2, calcium-dependent protein kinase 2, ATCPK11, CDPK2, calcium-dependent protein kinase 2, CPK11 |
-0.244 |
0.109 |
| 175 |
248801_at |
AT5G47370
|
HAT2 |
-0.242 |
-0.131 |
| 176 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.240 |
0.216 |
| 177 |
253165_at |
AT4G35320
|
unknown |
-0.239 |
-0.069 |
| 178 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
-0.237 |
0.021 |
| 179 |
256671_at |
AT3G52290
|
IQD3, IQ-domain 3 |
-0.235 |
-0.026 |
| 180 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.234 |
-0.018 |
| 181 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.232 |
-0.134 |
| 182 |
254382_at |
AT4G21890
|
unknown |
-0.231 |
-0.012 |
| 183 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.231 |
-0.073 |