|
probeID |
AGICode |
Annotation |
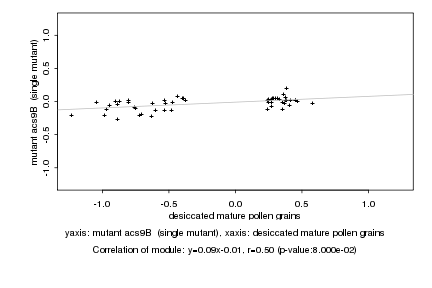
Log2 signal ratio
desiccated mature pollen grains |
Log2 signal ratio
mutant acs9B (single mutant) |
| 1 |
250915_at |
AT5G03790
|
ATHB51, HB51, homeobox 51, LMI1, LATE MERISTEM IDENTITY1 |
0.574 |
-0.021 |
| 2 |
262757_at |
AT1G16380
|
ATCHX1, CHX1, CATION EXCHANGER 1 |
0.466 |
0.005 |
| 3 |
252380_at |
AT3G47740
|
ABCA3, ATP-binding cassette A3, ATATH2, A. THALIANA ABC2 HOMOLOG 2, ATH2, ABC2 homolog 2 |
0.448 |
0.017 |
| 4 |
258272_at |
AT3G15610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.447 |
0.016 |
| 5 |
256029_at |
AT1G34130
|
STT3B, staurosporin and temperature sensitive 3-like b |
0.410 |
0.020 |
| 6 |
249427_at |
AT5G39850
|
[Ribosomal protein S4] |
0.399 |
-0.050 |
| 7 |
258541_at |
AT3G07000
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.383 |
0.202 |
| 8 |
266705_at |
AT2G19750
|
[Ribosomal protein S30 family protein] |
0.379 |
0.017 |
| 9 |
258504_at |
AT3G02590
|
[Fatty acid hydroxylase superfamily protein] |
0.373 |
0.025 |
| 10 |
260134_at |
AT1G66370
|
AtMYB113, myb domain protein 113, MYB113, myb domain protein 113 |
0.369 |
0.072 |
| 11 |
255793_at |
AT2G33250
|
unknown |
0.363 |
-0.029 |
| 12 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
0.354 |
0.109 |
| 13 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.352 |
-0.120 |
| 14 |
264462_at |
AT1G10200
|
AtWLIM1, WLIM1, WLIM1 |
0.351 |
-0.003 |
| 15 |
256780_at |
AT3G13640
|
ABCE1, ATP-binding cassette E1, ATRLI1, RNAse l inhibitor protein 1, RLI1, RNAse l inhibitor protein 1 |
0.327 |
0.033 |
| 16 |
251439_at |
AT3G59950
|
[Peptidase family C54 protein] |
0.314 |
0.047 |
| 17 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.294 |
0.056 |
| 18 |
251512_at |
AT3G59190
|
[F-box/RNI-like superfamily protein] |
0.281 |
0.058 |
| 19 |
245961_at |
AT5G19670
|
[Exostosin family protein] |
0.276 |
0.051 |
| 20 |
254351_at |
AT4G22300
|
SOBER1, SUPPRESSOR OF AVRBST-ELICITED RESISTANCE 1 |
0.271 |
-0.012 |
| 21 |
259061_at |
AT3G07410
|
AtRABA5b, RAB GTPase homolog A5B, RABA5b, RAB GTPase homolog A5B |
0.270 |
-0.061 |
| 22 |
245710_at |
AT5G04330
|
CYP84A4, CYTOCHROME P450 84A4 |
0.269 |
0.035 |
| 23 |
248215_at |
AT5G53680
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.248 |
0.020 |
| 24 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
0.247 |
0.022 |
| 25 |
254002_at |
AT4G26280
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.245 |
-0.009 |
| 26 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
0.242 |
0.045 |
| 27 |
257198_at |
AT3G23690
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.239 |
0.022 |
| 28 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.239 |
-0.109 |
| 29 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-1.238 |
-0.200 |
| 30 |
263095_at |
AT2G16040
|
[hAT dimerisation domain-containing protein / transposase-related] |
-1.048 |
-0.014 |
| 31 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.987 |
-0.204 |
| 32 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.976 |
-0.110 |
| 33 |
248045_at |
AT5G56030
|
AtHsp90.2, HEAT SHOCK PROTEIN 90.2, ERD8, EARLY-RESPONSIVE TO DEHYDRATION 8, HSP81-2, heat shock protein 81-2, HSP81.2, heat shock protein 81.2, HSP90.2, HEAT SHOCK PROTEIN 90.2 |
-0.953 |
-0.057 |
| 34 |
246096_at |
AT5G20330
|
BETAG4, beta-1,3-glucanase 4 |
-0.902 |
0.013 |
| 35 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-0.890 |
-0.036 |
| 36 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.888 |
-0.266 |
| 37 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
-0.879 |
0.005 |
| 38 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
-0.809 |
-0.014 |
| 39 |
258330_at |
AT3G16130
|
ATROPGEF13, PIRF2, phytochrome interacting RopGEF 2, ROPGEF13, ROP (rho of plants) guanine nucleotide exchange factor 13 |
-0.806 |
0.018 |
| 40 |
257822_at |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
-0.765 |
-0.088 |
| 41 |
250858_at |
AT5G04760
|
[Duplicated homeodomain-like superfamily protein] |
-0.752 |
-0.094 |
| 42 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.722 |
-0.197 |
| 43 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
-0.713 |
-0.191 |
| 44 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-0.632 |
-0.219 |
| 45 |
263613_at |
AT2G25250
|
unknown |
-0.626 |
-0.016 |
| 46 |
252278_at |
AT3G49530
|
ANAC062, NAC domain containing protein 62, NAC062, NAC domain containing protein 62, NTL6, NTM1 (NAC WITH TRANSMEMBRANE MOTIF 1)-LIKE 6 |
-0.603 |
-0.130 |
| 47 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
-0.540 |
-0.126 |
| 48 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
-0.540 |
0.028 |
| 49 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.529 |
-0.020 |
| 50 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.486 |
-0.122 |
| 51 |
259064_at |
AT3G07490
|
AGD11, ARF-GAP domain 11, AtCML3, CML3, calmodulin-like 3 |
-0.475 |
-0.010 |
| 52 |
249604_x_at |
AT5G37230
|
[RING/U-box superfamily protein] |
-0.441 |
0.079 |
| 53 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
-0.402 |
0.048 |
| 54 |
255235_at |
AT4G05510
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 4.0e-144 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
-0.391 |
0.057 |
| 55 |
245026_at |
ATCG00140
|
ATPH |
-0.378 |
0.019 |