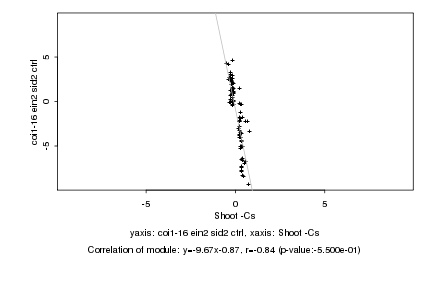
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Shoot -Cs |
Log2 signal ratio
coi1-16 ein2 sid2 ctrl |
| 1 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.761 |
-3.365 |
| 2 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.704 |
-9.217 |
| 3 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.674 |
-2.224 |
| 4 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.538 |
-2.183 |
| 5 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.516 |
-6.687 |
| 6 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.501 |
-6.904 |
| 7 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.429 |
-8.379 |
| 8 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.391 |
-4.950 |
| 9 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.389 |
-8.292 |
| 10 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.376 |
-6.572 |
| 11 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.345 |
-6.357 |
| 12 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.342 |
-1.689 |
| 13 |
263515_at |
AT2G21640
|
unknown |
0.327 |
-0.327 |
| 14 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.326 |
-7.768 |
| 15 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.311 |
-3.539 |
| 16 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.306 |
-4.987 |
| 17 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.300 |
-7.409 |
| 18 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.296 |
-4.421 |
| 19 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.293 |
-4.271 |
| 20 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.292 |
-6.493 |
| 21 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.291 |
-7.201 |
| 22 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.289 |
-7.739 |
| 23 |
249252_at |
AT5G42010
|
[Transducin/WD40 repeat-like superfamily protein] |
0.281 |
-5.222 |
| 24 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.281 |
-3.421 |
| 25 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.279 |
-5.002 |
| 26 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.275 |
-1.801 |
| 27 |
249515_at |
AT5G38530
|
TSBtype2, tryptophan synthase beta type 2 |
0.254 |
-0.268 |
| 28 |
256883_at |
AT3G26440
|
unknown |
0.247 |
-3.943 |
| 29 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.244 |
-1.144 |
| 30 |
266536_at |
AT2G16900
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.217 |
-3.730 |
| 31 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.211 |
-2.162 |
| 32 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.205 |
-3.255 |
| 33 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
0.204 |
-2.063 |
| 34 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.192 |
-2.230 |
| 35 |
253224_at |
AT4G34860
|
A/N-InvB, alkaline/neutral invertase B |
0.190 |
-0.180 |
| 36 |
250738_at |
AT5G05730
|
AMT1, A-METHYL TRYPTOPHAN RESISTANT 1, ASA1, anthranilate synthase alpha subunit 1, JDL1, JASMONATE-INDUCED DEFECTIVE LATERAL ROOT 1, TRP5, TRYPTOPHAN BIOSYNTHESIS 5, WEI2, WEAK ETHYLENE INSENSITIVE 2 |
0.187 |
-3.969 |
| 37 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
0.187 |
-2.754 |
| 38 |
246042_at |
AT5G19440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.185 |
-1.725 |
| 39 |
264941_at |
AT1G60680
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.185 |
1.513 |
| 40 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
0.172 |
-3.638 |
| 41 |
249372_at |
AT5G40760
|
G6PD6, glucose-6-phosphate dehydrogenase 6 |
0.171 |
-2.938 |
| 42 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.530 |
4.332 |
| 43 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.406 |
2.571 |
| 44 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.397 |
4.167 |
| 45 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.366 |
-0.066 |
| 46 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.342 |
2.761 |
| 47 |
252765_at |
AT3G42800
|
unknown |
-0.329 |
3.358 |
| 48 |
257858_at |
AT3G12920
|
BRG3, BOI-related gene 3 |
-0.306 |
-0.030 |
| 49 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.304 |
3.092 |
| 50 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.300 |
2.745 |
| 51 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.293 |
2.390 |
| 52 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.293 |
1.343 |
| 53 |
254787_at |
AT4G12690
|
unknown |
-0.292 |
0.740 |
| 54 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.289 |
0.259 |
| 55 |
246952_at |
AT5G04820
|
ATOFP13, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 13, OFP13, ovate family protein 13 |
-0.272 |
0.849 |
| 56 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.237 |
1.932 |
| 57 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.232 |
2.431 |
| 58 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
-0.223 |
1.337 |
| 59 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.222 |
2.266 |
| 60 |
257986_at |
AT3G20865
|
AGP40, arabinogalactan protein 40 |
-0.211 |
-0.424 |
| 61 |
256066_at |
AT1G06980
|
unknown |
-0.205 |
4.710 |
| 62 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.196 |
1.095 |
| 63 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.194 |
2.967 |
| 64 |
245176_at |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.191 |
1.625 |
| 65 |
249355_at |
AT5G40500
|
unknown |
-0.190 |
0.685 |
| 66 |
263305_at |
AT2G01930
|
ATBPC1, BASIC PENTACYSTEINE1, BBR, BPC1, basic pentacysteine1 |
-0.177 |
-0.329 |
| 67 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
-0.176 |
0.024 |
| 68 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
-0.175 |
0.959 |
| 69 |
264096_at |
AT1G78995
|
unknown |
-0.170 |
2.157 |
| 70 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.167 |
1.514 |
| 71 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
-0.167 |
1.381 |
| 72 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.166 |
0.528 |
| 73 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.165 |
2.589 |
| 74 |
258676_at |
AT3G08600
|
unknown |
-0.164 |
2.058 |
| 75 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.164 |
1.552 |
| 76 |
261742_at |
AT1G08390
|
unknown |
-0.163 |
1.124 |
| 77 |
257811_at |
AT3G25280
|
AtNPF4.2, NPF4.2, NRT1/ PTR family 4.2 |
-0.163 |
0.099 |
| 78 |
253679_at |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
-0.162 |
0.982 |
| 79 |
254303_at |
AT4G22830
|
unknown |
-0.162 |
0.185 |
| 80 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.157 |
1.226 |
| 81 |
255878_at |
AT2G40620
|
[Basic-leucine zipper (bZIP) transcription factor family protein] |
-0.157 |
-0.301 |