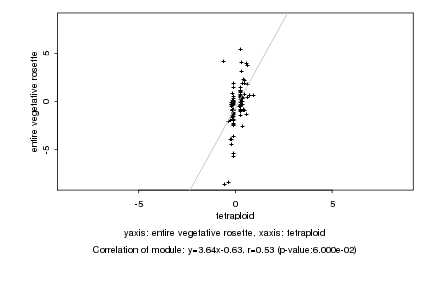
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
tetraploid |
Log2 signal ratio
entire vegetative rosette |
| 1 |
262211_at |
AT1G74930
|
ORA47 |
0.882 |
0.680 |
| 2 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.694 |
0.724 |
| 3 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.598 |
0.472 |
| 4 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.596 |
1.854 |
| 5 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.580 |
3.748 |
| 6 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.560 |
-1.257 |
| 7 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.553 |
4.021 |
| 8 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.460 |
1.922 |
| 9 |
253643_at |
AT4G29780
|
unknown |
0.457 |
-0.887 |
| 10 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.457 |
2.251 |
| 11 |
245041_at |
AT2G26530
|
AR781 |
0.421 |
0.809 |
| 12 |
265184_at |
AT1G23710
|
unknown |
0.387 |
-0.836 |
| 13 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.378 |
2.299 |
| 14 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.372 |
-0.793 |
| 15 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.359 |
0.348 |
| 16 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.352 |
1.921 |
| 17 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.350 |
-0.297 |
| 18 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.346 |
0.065 |
| 19 |
265387_at |
AT2G20670
|
unknown |
0.345 |
0.481 |
| 20 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.338 |
-2.515 |
| 21 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.324 |
0.408 |
| 22 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.309 |
3.122 |
| 23 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.298 |
0.522 |
| 24 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.288 |
0.099 |
| 25 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.287 |
0.318 |
| 26 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.276 |
-0.291 |
| 27 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.266 |
4.081 |
| 28 |
263265_at |
AT2G38820
|
unknown |
0.252 |
1.159 |
| 29 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.249 |
0.684 |
| 30 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.248 |
-0.089 |
| 31 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.247 |
5.445 |
| 32 |
259979_at |
AT1G76600
|
unknown |
0.247 |
-0.792 |
| 33 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.245 |
-1.033 |
| 34 |
247585_at |
AT5G60680
|
unknown |
0.240 |
0.996 |
| 35 |
266259_at |
AT2G27830
|
unknown |
0.237 |
1.554 |
| 36 |
247708_at |
AT5G59550
|
unknown |
0.236 |
-0.096 |
| 37 |
264379_at |
AT2G25200
|
unknown |
0.235 |
0.788 |
| 38 |
264210_at |
AT1G22640
|
ATMYB3, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 3, MYB3, myb domain protein 3 |
0.233 |
-0.459 |
| 39 |
259015_at |
AT3G07350
|
unknown |
0.230 |
0.564 |
| 40 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.223 |
0.002 |
| 41 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
0.220 |
0.313 |
| 42 |
259231_at |
AT3G11410
|
AHG3, ATPP2CA, ARABIDOPSIS THALIANA PROTEIN PHOSPHATASE 2CA, PP2CA, protein phosphatase 2CA |
0.219 |
1.095 |
| 43 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.218 |
-0.922 |
| 44 |
253322_at |
AT4G33980
|
unknown |
0.217 |
-1.361 |
| 45 |
246289_at |
AT3G56880
|
[VQ motif-containing protein] |
0.217 |
0.430 |
| 46 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.215 |
-0.275 |
| 47 |
262135_at |
AT1G78080
|
AtWIND1, RAP2.4, related to AP2 4, WIND1, wound induced dedifferentiation 1 |
0.214 |
-0.371 |
| 48 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.211 |
1.190 |
| 49 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.206 |
-0.463 |
| 50 |
254574_at |
AT4G19430
|
unknown |
-0.666 |
4.243 |
| 51 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.583 |
-8.512 |
| 52 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
-0.403 |
-8.337 |
| 53 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.361 |
-1.980 |
| 54 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
-0.296 |
-1.902 |
| 55 |
257421_at |
AT1G12030
|
unknown |
-0.264 |
-3.918 |
| 56 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.252 |
-3.923 |
| 57 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.241 |
-0.234 |
| 58 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.234 |
-4.440 |
| 59 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.207 |
-0.272 |
| 60 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
-0.207 |
-0.099 |
| 61 |
252661_at |
AT3G44450
|
unknown |
-0.205 |
-0.413 |
| 62 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.200 |
-1.481 |
| 63 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.194 |
-1.440 |
| 64 |
258677_at |
AT3G08730
|
ATPK1, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 1, ATPK6, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 6, ATS6K1, PK1, protein-serine kinase 1, PK6, ROTEIN-SERINE KINASE 6, S6K1, P70 RIBOSOMAL S6 KINASE |
-0.192 |
-0.799 |
| 65 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.191 |
-0.363 |
| 66 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
-0.191 |
-0.385 |
| 67 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.183 |
-0.865 |
| 68 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.176 |
-1.807 |
| 69 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
-0.175 |
-0.185 |
| 70 |
263808_at |
AT2G04340
|
unknown |
-0.173 |
0.160 |
| 71 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-0.169 |
-0.894 |
| 72 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.169 |
-1.605 |
| 73 |
257076_at |
AT3G19680
|
unknown |
-0.155 |
0.908 |
| 74 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.151 |
-1.285 |
| 75 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-0.150 |
-2.470 |
| 76 |
248028_at |
AT5G55620
|
unknown |
-0.150 |
-1.511 |
| 77 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
-0.147 |
-3.613 |
| 78 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.146 |
0.402 |
| 79 |
257181_at |
AT3G13190
|
unknown |
-0.145 |
-0.089 |
| 80 |
245692_at |
AT5G04150
|
BHLH101 |
-0.138 |
-5.608 |
| 81 |
262328_at |
AT1G64100
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.136 |
1.939 |
| 82 |
260951_at |
AT1G06150
|
EMB1444, EMBRYO DEFECTIVE 1444 |
-0.136 |
-0.231 |
| 83 |
259968_at |
AT1G76530
|
[Auxin efflux carrier family protein] |
-0.135 |
-1.118 |
| 84 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
-0.135 |
0.142 |
| 85 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
-0.135 |
-5.366 |
| 86 |
253940_at |
AT4G26950
|
unknown |
-0.134 |
-2.231 |
| 87 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.131 |
0.091 |
| 88 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.130 |
-1.846 |
| 89 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
-0.128 |
-1.888 |
| 90 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
-0.126 |
1.476 |
| 91 |
263873_at |
AT2G21860
|
[violaxanthin de-epoxidase-related] |
-0.126 |
-0.172 |
| 92 |
262446_at |
AT1G49310
|
unknown |
-0.125 |
-2.359 |
| 93 |
258009_at |
AT3G19440
|
[Pseudouridine synthase family protein] |
-0.123 |
-1.196 |
| 94 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
-0.123 |
-0.771 |
| 95 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.122 |
0.056 |
| 96 |
256923_at |
AT3G29635
|
[HXXXD-type acyl-transferase family protein] |
-0.121 |
-0.851 |
| 97 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
-0.120 |
0.565 |