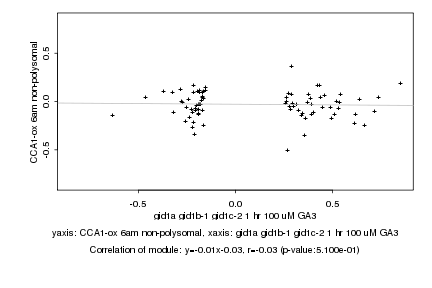
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gid1a gid1b-1 gid1c-2 1 hr 100 uM GA3 |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.849 |
0.194 |
| 2 |
261866_at |
AT1G50420
|
SCL-3, SCARECROW-LIKE 3, SCL3, scarecrow-like 3 |
0.734 |
0.047 |
| 3 |
265891_at |
AT2G15010
|
[Plant thionin] |
0.712 |
-0.099 |
| 4 |
266912_at |
AT2G45900
|
TRM13, TON1 Recruiting Motif 13 |
0.661 |
-0.244 |
| 5 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.638 |
0.021 |
| 6 |
250611_at |
AT5G07200
|
ATGA20OX3, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 3, GA20OX3, gibberellin 20-oxidase 3, YAP169 |
0.618 |
-0.127 |
| 7 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
0.611 |
-0.219 |
| 8 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.541 |
0.073 |
| 9 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.533 |
-0.003 |
| 10 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.528 |
-0.069 |
| 11 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.521 |
0.001 |
| 12 |
247061_at |
AT5G66780
|
unknown |
0.509 |
-0.128 |
| 13 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
0.494 |
-0.171 |
| 14 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.489 |
-0.061 |
| 15 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.456 |
0.064 |
| 16 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
0.448 |
-0.059 |
| 17 |
260883_at |
AT1G29270
|
unknown |
0.437 |
0.046 |
| 18 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.429 |
0.176 |
| 19 |
245341_at |
AT4G16447
|
unknown |
0.423 |
0.171 |
| 20 |
255900_at |
AT1G17830
|
unknown |
0.402 |
-0.104 |
| 21 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.388 |
-0.131 |
| 22 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.387 |
-0.028 |
| 23 |
246929_at |
AT5G25210
|
unknown |
0.385 |
0.037 |
| 24 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.376 |
0.074 |
| 25 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.371 |
-0.009 |
| 26 |
263829_at |
AT2G40435
|
unknown |
0.357 |
-0.167 |
| 27 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.352 |
-0.346 |
| 28 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.344 |
-0.118 |
| 29 |
263325_at |
AT2G04240
|
XERICO |
0.339 |
-0.137 |
| 30 |
266124_at |
AT2G45080
|
cycp3;1, cyclin p3;1 |
0.324 |
-0.083 |
| 31 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.310 |
-0.025 |
| 32 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.295 |
-0.050 |
| 33 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.290 |
-0.018 |
| 34 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
0.287 |
0.083 |
| 35 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.285 |
0.370 |
| 36 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.282 |
-0.077 |
| 37 |
247212_at |
AT5G65040
|
unknown |
0.276 |
-0.049 |
| 38 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
0.272 |
0.086 |
| 39 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.266 |
-0.505 |
| 40 |
253774_at |
AT4G28530
|
anac074, NAC domain containing protein 74, NAC074, NAC domain containing protein 74 |
0.262 |
0.008 |
| 41 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.261 |
0.046 |
| 42 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
0.256 |
-0.019 |
| 43 |
255461_at |
AT4G02780
|
ABC33, ATCPS1, ARABIDOPSIS THALIANA ENT-COPALYL DIPHOSPHATE SYNTHETASE 1, CPS, CPP synthase, CPS1, ENT-COPALYL DIPHOSPHATE SYNTHETASE 1, GA1, GA REQUIRING 1 |
-0.634 |
-0.141 |
| 44 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
-0.467 |
0.049 |
| 45 |
258091_at |
AT3G14560
|
unknown |
-0.372 |
0.112 |
| 46 |
247474_at |
AT5G62280
|
unknown |
-0.326 |
0.102 |
| 47 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.320 |
-0.106 |
| 48 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.288 |
0.133 |
| 49 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.280 |
0.002 |
| 50 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
-0.273 |
-0.001 |
| 51 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.259 |
-0.201 |
| 52 |
247177_at |
AT5G65300
|
unknown |
-0.255 |
-0.055 |
| 53 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.245 |
0.030 |
| 54 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.237 |
-0.156 |
| 55 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.231 |
-0.073 |
| 56 |
267515_at |
AT2G45680
|
TCP9, TCP domain protein 9 |
-0.226 |
-0.260 |
| 57 |
262850_at |
AT1G14920
|
GAI, GIBBERELLIC ACID INSENSITIVE, RGA2, RESTORATION ON GROWTH ON AMMONIA 2 |
-0.223 |
-0.104 |
| 58 |
250327_at |
AT5G12050
|
unknown |
-0.219 |
0.173 |
| 59 |
256720_at |
AT2G34140
|
[Dof-type zinc finger DNA-binding family protein] |
-0.219 |
-0.217 |
| 60 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.217 |
0.095 |
| 61 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.215 |
-0.340 |
| 62 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.210 |
-0.065 |
| 63 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
-0.208 |
-0.091 |
| 64 |
259985_at |
AT1G76620
|
unknown |
-0.203 |
-0.036 |
| 65 |
249211_at |
AT5G42680
|
unknown |
-0.197 |
0.106 |
| 66 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.194 |
-0.078 |
| 67 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.194 |
-0.026 |
| 68 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.194 |
-0.118 |
| 69 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.191 |
-0.125 |
| 70 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.189 |
-0.027 |
| 71 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.188 |
0.103 |
| 72 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.188 |
0.123 |
| 73 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.182 |
-0.021 |
| 74 |
252129_at |
AT3G50890
|
AtHB28, homeobox protein 28, HB28, homeobox protein 28, ZHD7, ZINC FINGER HOMEODOMAIN 7 |
-0.176 |
0.016 |
| 75 |
260081_at |
AT1G78170
|
unknown |
-0.173 |
0.061 |
| 76 |
250569_at |
AT5G08130
|
BIM1 |
-0.173 |
0.050 |
| 77 |
260226_at |
AT1G74660
|
MIF1, mini zinc finger 1 |
-0.172 |
-0.083 |
| 78 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-0.171 |
0.099 |
| 79 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.168 |
0.106 |
| 80 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.168 |
0.035 |
| 81 |
251601_at |
AT3G57800
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.167 |
-0.239 |
| 82 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.159 |
0.121 |
| 83 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.158 |
0.150 |