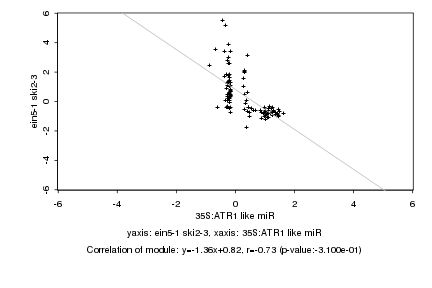
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S:ATR1 like miR |
Log2 signal ratio
ein5-1 ski2-3 |
| 1 |
245013_at |
ATCG00470
|
ATPE, ATP synthase epsilon chain |
1.601 |
-0.805 |
| 2 |
244935_at |
ATCG01090
|
NDHI |
1.480 |
-0.672 |
| 3 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
1.454 |
-1.012 |
| 4 |
244969_at |
ATCG00650
|
RPS18, ribosomal protein S18 |
1.449 |
-0.496 |
| 5 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
1.448 |
-0.903 |
| 6 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
1.421 |
-0.760 |
| 7 |
244984_at |
ATCG00800
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
1.399 |
-0.871 |
| 8 |
244962_at |
ATCG01050
|
NDHD |
1.347 |
-0.735 |
| 9 |
244971_at |
ATCG00670
|
CLPP1, CASEINOLYTIC PROTEASE P 1, PCLPP, plastid-encoded CLP P |
1.326 |
-0.853 |
| 10 |
245006_at |
ATCG00340
|
PSAB |
1.271 |
-0.612 |
| 11 |
244965_at |
ATCG00590
|
ORF31 |
1.258 |
-0.577 |
| 12 |
244976_at |
ATCG00720
|
PETB, photosynthetic electron transfer B |
1.258 |
-0.669 |
| 13 |
245014_at |
ATCG00480
|
ATPB, ATP synthase subunit beta, PB, ATP synthase subunit beta |
1.243 |
-0.729 |
| 14 |
245025_at |
ATCG00130
|
ATPF |
1.238 |
-0.899 |
| 15 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
1.230 |
-0.404 |
| 16 |
245016_at |
ATCG00500
|
ACCD, acetyl-CoA carboxylase carboxyl transferase subunit beta |
1.204 |
-0.411 |
| 17 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
1.165 |
-0.767 |
| 18 |
244968_at |
ATCG00640
|
RPL33, ribosomal protein L33 |
1.126 |
-0.311 |
| 19 |
245001_at |
ATCG00220
|
PSBM, photosystem II reaction center protein M |
1.109 |
-0.429 |
| 20 |
244932_at |
ATCG01060
|
PSAC |
1.095 |
-0.780 |
| 21 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
1.091 |
-1.045 |
| 22 |
245021_at |
ATCG00550
|
PSBJ, photosystem II reaction center protein J |
1.091 |
-0.589 |
| 23 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
1.061 |
-1.057 |
| 24 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
1.052 |
-0.749 |
| 25 |
244998_at |
ATCG00180
|
RPOC1 |
1.030 |
-0.821 |
| 26 |
244997_at |
ATCG00170
|
RPOC2 |
1.003 |
-0.946 |
| 27 |
244964_at |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
0.996 |
-0.605 |
| 28 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
0.989 |
-1.159 |
| 29 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
0.984 |
-0.654 |
| 30 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
0.976 |
-0.711 |
| 31 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
0.954 |
-0.971 |
| 32 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
0.953 |
-0.397 |
| 33 |
245020_at |
ATCG00540
|
PETA, photosynthetic electron transfer A |
0.918 |
-0.747 |
| 34 |
245004_at |
ATCG00300
|
YCF9 |
0.881 |
-0.683 |
| 35 |
244938_at |
ATCG01120
|
RPS15, chloroplast ribosomal protein S15 |
0.867 |
-1.134 |
| 36 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
0.818 |
-0.559 |
| 37 |
245023_at |
ATCG00080
|
PSBI, photosystem II reaction center protein I |
0.657 |
-0.596 |
| 38 |
245012_at |
ATCG00440
|
NDHC |
0.611 |
-0.563 |
| 39 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.518 |
-0.434 |
| 40 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.460 |
-0.743 |
| 41 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.453 |
-0.982 |
| 42 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.435 |
-0.406 |
| 43 |
254954_at |
AT4G10910
|
unknown |
0.428 |
-0.406 |
| 44 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.380 |
-0.674 |
| 45 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.379 |
3.151 |
| 46 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.379 |
0.667 |
| 47 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
0.372 |
-1.742 |
| 48 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.372 |
0.106 |
| 49 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
0.314 |
-0.108 |
| 50 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
0.304 |
0.518 |
| 51 |
247149_at |
AT5G65660
|
[hydroxyproline-rich glycoprotein family protein] |
0.295 |
1.995 |
| 52 |
260367_at |
AT1G69760
|
unknown |
0.292 |
2.153 |
| 53 |
251700_at |
AT3G56640
|
SEC15A, exocyst complex component sec15A |
0.289 |
-0.517 |
| 54 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
0.277 |
2.061 |
| 55 |
253476_at |
AT4G32300
|
SD2-5, S-domain-2 5 |
0.268 |
1.628 |
| 56 |
245176_at |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.266 |
1.027 |
| 57 |
261684_at |
AT1G47400
|
unknown |
-0.898 |
2.483 |
| 58 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-0.702 |
3.581 |
| 59 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.616 |
-0.365 |
| 60 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
-0.451 |
5.574 |
| 61 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
-0.400 |
3.427 |
| 62 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.384 |
1.714 |
| 63 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.359 |
5.210 |
| 64 |
249656_at |
AT5G37130
|
[Protein prenylyltransferase superfamily protein] |
-0.354 |
0.127 |
| 65 |
245090_at |
AT2G40900
|
UMAMIT11, Usually multiple acids move in and out Transporters 11 |
-0.331 |
1.859 |
| 66 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.324 |
-0.373 |
| 67 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
-0.303 |
0.901 |
| 68 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
-0.294 |
2.795 |
| 69 |
262322_at |
AT1G27590
|
unknown |
-0.293 |
0.372 |
| 70 |
250487_at |
AT5G09690
|
ATMGT7, ARABIDOPSIS THALIANA MAGNESIUM TRANSPORTER 7, MGT7, magnesium transporter 7, MRS2-7 |
-0.290 |
0.119 |
| 71 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.289 |
-0.301 |
| 72 |
252628_at |
AT3G44960
|
unknown |
-0.277 |
-0.406 |
| 73 |
255710_at |
AT4G00030
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
-0.276 |
1.257 |
| 74 |
264270_at |
AT1G60260
|
BGLU5, beta glucosidase 5 |
-0.273 |
0.540 |
| 75 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.272 |
1.340 |
| 76 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.269 |
1.143 |
| 77 |
263478_at |
AT2G31880
|
EVR, EVERSHED, SOBIR1, SUPPRESSOR OF BIR1 1 |
-0.264 |
3.029 |
| 78 |
254784_at |
AT4G12720
|
AtNUDT7, Arabidopsis thaliana Nudix hydrolase homolog 7, GFG1, GROWTH FACTOR GENE 1, NUDT7 |
-0.263 |
3.882 |
| 79 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
-0.259 |
1.786 |
| 80 |
247854_at |
AT5G58200
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.254 |
0.270 |
| 81 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.239 |
1.394 |
| 82 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.237 |
2.629 |
| 83 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.236 |
0.256 |
| 84 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.234 |
0.556 |
| 85 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
-0.233 |
-0.425 |
| 86 |
253913_at |
AT4G27370
|
ATVIIIB, MYOSIN VIII B, VIIIB |
-0.231 |
-0.038 |
| 87 |
251594_at |
AT3G57630
|
[exostosin family protein] |
-0.228 |
1.059 |
| 88 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.225 |
1.657 |
| 89 |
259611_at |
AT1G52280
|
AtRABG3d, RAB GTPase homolog G3D, RABG3d, RAB GTPase homolog G3D |
-0.225 |
0.322 |
| 90 |
265898_at |
AT2G25690
|
unknown |
-0.223 |
2.601 |
| 91 |
266222_at |
AT2G28780
|
unknown |
-0.222 |
0.256 |
| 92 |
265357_at |
AT2G16740
|
UBC29, ubiquitin-conjugating enzyme 29 |
-0.215 |
-0.039 |
| 93 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.210 |
0.130 |
| 94 |
265731_at |
AT2G01130
|
[DEA(D/H)-box RNA helicase family protein] |
-0.208 |
0.656 |
| 95 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
-0.207 |
1.862 |
| 96 |
259034_at |
AT3G09410
|
[Pectinacetylesterase family protein] |
-0.205 |
0.362 |
| 97 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
-0.204 |
0.392 |
| 98 |
251968_at |
AT3G53100
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.202 |
1.462 |
| 99 |
250186_at |
AT5G14500
|
[aldose 1-epimerase family protein] |
-0.199 |
0.362 |
| 100 |
255529_at |
AT4G02120
|
[CTP synthase family protein] |
-0.196 |
0.428 |
| 101 |
262328_at |
AT1G64100
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.195 |
-0.394 |
| 102 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.193 |
1.312 |
| 103 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.193 |
1.109 |
| 104 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.192 |
0.746 |
| 105 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.189 |
0.837 |
| 106 |
259380_at |
AT3G16340
|
ABCG29, ATP-binding cassette G29, AtABCG29, ATPDR1, PLEIOTROPIC DRUG RESISTANCE 1, PDR1, pleiotropic drug resistance 1 |
-0.189 |
0.505 |
| 107 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
-0.188 |
3.402 |
| 108 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.187 |
-0.719 |
| 109 |
251903_at |
AT3G54120
|
[Reticulon family protein] |
-0.187 |
0.769 |
| 110 |
261294_at |
AT1G48430
|
[Dihydroxyacetone kinase] |
-0.185 |
0.453 |
| 111 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
-0.185 |
0.708 |