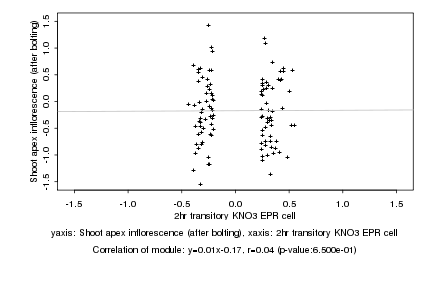
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
2hr transitory KNO3 EPR cell |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.548 |
-0.436 |
| 2 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.523 |
0.597 |
| 3 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
0.520 |
-0.448 |
| 4 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.503 |
0.188 |
| 5 |
260522_x_at |
AT2G41730
|
unknown |
0.482 |
-1.036 |
| 6 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
0.444 |
0.576 |
| 7 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
0.441 |
0.631 |
| 8 |
256703_at |
AT3G30260
|
AGL79, AGAMOUS-like 79 |
0.433 |
-0.127 |
| 9 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.420 |
0.414 |
| 10 |
262399_at |
AT1G49500
|
unknown |
0.415 |
0.569 |
| 11 |
258671_at |
AT3G08560
|
VHA-E2, vacuolar H+-ATPase subunit E isoform 2 |
0.412 |
0.396 |
| 12 |
250472_at |
AT5G10210
|
unknown |
0.406 |
-0.944 |
| 13 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.397 |
0.417 |
| 14 |
253653_at |
AT4G30050
|
unknown |
0.382 |
-0.730 |
| 15 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.368 |
-0.863 |
| 16 |
265784_at |
AT2G07280
|
unknown |
0.346 |
-0.964 |
| 17 |
249377_at |
AT5G40690
|
unknown |
0.345 |
-0.181 |
| 18 |
250201_at |
AT5G14230
|
unknown |
0.344 |
0.736 |
| 19 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
0.343 |
0.258 |
| 20 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
0.332 |
-0.344 |
| 21 |
248925_at |
AT5G45910
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.330 |
-0.447 |
| 22 |
265984_at |
AT2G24210
|
TPS10, terpene synthase 10 |
0.327 |
-0.845 |
| 23 |
253939_at |
AT4G26930
|
AtMYB97, myb domain protein 97, MYB97, myb domain protein 97 |
0.325 |
-1.359 |
| 24 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
0.325 |
-0.287 |
| 25 |
249896_at |
AT5G22530
|
unknown |
0.323 |
-0.643 |
| 26 |
261737_at |
AT1G47885
|
[Ribonuclease inhibitor] |
0.319 |
-0.744 |
| 27 |
248289_at |
AT5G52880
|
[F-box family protein] |
0.311 |
-0.355 |
| 28 |
253990_at |
AT4G26160
|
ACHT1, atypical CYS HIS rich thioredoxin 1 |
0.301 |
-0.151 |
| 29 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.299 |
0.318 |
| 30 |
253322_at |
AT4G33980
|
unknown |
0.293 |
-0.998 |
| 31 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
0.293 |
-0.307 |
| 32 |
265602_at |
AT2G14380
|
[gypsy-like retrotransposon family, has a 1.6e-123 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.291 |
-0.382 |
| 33 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
0.289 |
0.361 |
| 34 |
257851_at |
AT3G12940
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.288 |
0.244 |
| 35 |
258959_at |
AT3G10600
|
CAT7, cationic amino acid transporter 7 |
0.286 |
-0.034 |
| 36 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
0.278 |
1.102 |
| 37 |
253305_at |
AT4G33666
|
unknown |
0.276 |
-0.821 |
| 38 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.275 |
-0.475 |
| 39 |
257305_at |
AT3G30250
|
unknown |
0.274 |
-0.745 |
| 40 |
253049_at |
AT4G37300
|
MEE59, maternal effect embryo arrest 59 |
0.264 |
1.181 |
| 41 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.255 |
0.226 |
| 42 |
255125_at |
AT4G08250
|
[GRAS family transcription factor] |
0.252 |
-0.621 |
| 43 |
259363_at |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
0.252 |
0.418 |
| 44 |
259379_at |
AT3G16350
|
[Homeodomain-like superfamily protein] |
0.251 |
0.315 |
| 45 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
0.249 |
-0.263 |
| 46 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.249 |
-1.097 |
| 47 |
259213_at |
AT3G09010
|
[Protein kinase superfamily protein] |
0.247 |
-0.529 |
| 48 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.245 |
-1.013 |
| 49 |
262425_at |
AT1G47660
|
unknown |
0.245 |
0.115 |
| 50 |
256043_at |
AT1G07210
|
[Ribosomal protein S18] |
0.243 |
0.343 |
| 51 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
0.242 |
-0.144 |
| 52 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.242 |
-0.776 |
| 53 |
258974_at |
AT3G01890
|
[SWIB/MDM2 domain superfamily protein] |
0.242 |
-0.296 |
| 54 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.241 |
0.193 |
| 55 |
263642_at |
AT2G04750
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.235 |
-0.892 |
| 56 |
244974_at |
ATCG00700
|
PSBN, photosystem II reaction center protein N |
0.234 |
0.140 |
| 57 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.446 |
-0.040 |
| 58 |
248923_at |
AT5G45940
|
AtNUDT11, Arabidopsis thaliana nudix hydrolase homolog 11, AtNUDX11, Arabidopsis thaliana nudix hydrolase homolog 11, NUDT11, nudix hydrolase homolog 11, NUDX11, nudix hydrolase homolog 11 |
-0.396 |
0.686 |
| 59 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
-0.396 |
-1.282 |
| 60 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.389 |
-0.065 |
| 61 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
-0.378 |
-0.465 |
| 62 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.373 |
-0.956 |
| 63 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
-0.370 |
-0.792 |
| 64 |
257715_at |
AT3G12750
|
AtZIP1, ZIP1, zinc transporter 1 precursor |
-0.353 |
0.603 |
| 65 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
-0.351 |
-0.616 |
| 66 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.350 |
0.389 |
| 67 |
248212_at |
AT5G54020
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.348 |
-0.876 |
| 68 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.346 |
0.546 |
| 69 |
260395_at |
AT1G69780
|
ATHB13 |
-0.337 |
-0.006 |
| 70 |
258502_at |
AT3G02490
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.336 |
-0.371 |
| 71 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.333 |
0.631 |
| 72 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
-0.332 |
-0.452 |
| 73 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.330 |
-1.534 |
| 74 |
258134_at |
AT3G24530
|
[AAA-type ATPase family protein / ankyrin repeat family protein] |
-0.330 |
-0.311 |
| 75 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.326 |
-0.385 |
| 76 |
250395_at |
AT5G10950
|
[Tudor/PWWP/MBT superfamily protein] |
-0.317 |
-0.574 |
| 77 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.317 |
-0.200 |
| 78 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
-0.317 |
-0.798 |
| 79 |
265439_at |
AT2G21045
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.311 |
-0.754 |
| 80 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.308 |
-0.138 |
| 81 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
-0.307 |
0.451 |
| 82 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
-0.298 |
-0.502 |
| 83 |
258748_at |
AT3G05930
|
GLP8, germin-like protein 8 |
-0.286 |
-0.336 |
| 84 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.275 |
0.165 |
| 85 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-0.270 |
0.018 |
| 86 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
-0.269 |
0.421 |
| 87 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.263 |
0.285 |
| 88 |
260563_at |
AT2G43840
|
UGT74F1, UDP-glycosyltransferase 74 F1 |
-0.259 |
-1.178 |
| 89 |
256712_at |
AT2G34020
|
[Calcium-binding EF-hand family protein] |
-0.255 |
-1.041 |
| 90 |
262411_at |
AT1G34640
|
[peptidases] |
-0.253 |
1.428 |
| 91 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.249 |
-0.091 |
| 92 |
247059_at |
AT5G66690
|
UGT72E2 |
-0.248 |
-1.168 |
| 93 |
254281_at |
AT4G22840
|
[Sodium Bile acid symporter family] |
-0.244 |
0.581 |
| 94 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
-0.243 |
0.229 |
| 95 |
254971_at |
AT4G10380
|
AtNIP5;1, NIP5;1, NOD26-like intrinsic protein 5;1, NLM6, NOD26-LIKE MIP 6, NLM8, NOD26-LIKE MIP 8 |
-0.238 |
0.336 |
| 96 |
267632_at |
AT2G42160
|
BRIZ1, BRAP2 RING ZnF UBP domain-containing protein 1 |
-0.234 |
-0.276 |
| 97 |
256510_at |
AT1G33360
|
[ATP-dependent Clp protease] |
-0.234 |
-0.605 |
| 98 |
260639_at |
AT1G53180
|
unknown |
-0.232 |
0.157 |
| 99 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.231 |
-0.632 |
| 100 |
259162_at |
AT3G01640
|
ATGLCAK, ARABIDOPSIS THALIANA GLUCURONOKINASE, GLCAK, glucuronokinase G |
-0.230 |
-0.423 |
| 101 |
259849_at |
AT1G72190
|
[D-isomer specific 2-hydroxyacid dehydrogenase family protein] |
-0.227 |
0.588 |
| 102 |
261220_at |
AT1G19970
|
[ER lumen protein retaining receptor family protein] |
-0.226 |
1.016 |
| 103 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.226 |
-0.116 |
| 104 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.223 |
0.121 |
| 105 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
-0.223 |
0.937 |
| 106 |
266136_at |
AT2G45060
|
[Uncharacterised conserved protein UCP022280] |
-0.222 |
0.040 |
| 107 |
259351_at |
AT3G05150
|
[Major facilitator superfamily protein] |
-0.220 |
-0.160 |
| 108 |
253065_at |
AT4G37740
|
AtGRF2, growth-regulating factor 2, GRF2, growth-regulating factor 2 |
-0.217 |
-0.316 |
| 109 |
260930_at |
AT1G02620
|
[Ras-related small GTP-binding family protein] |
-0.210 |
-0.521 |
| 110 |
253605_at |
AT4G30990
|
[ARM repeat superfamily protein] |
-0.209 |
-0.246 |
| 111 |
245338_at |
AT4G16442
|
[Uncharacterised protein family (UPF0497)] |
-0.207 |
0.029 |