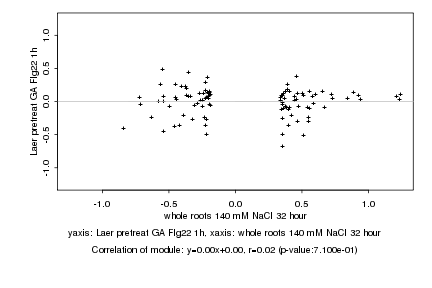
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
whole roots 140 mM NaCl 32 hour |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.238 |
0.121 |
| 2 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.233 |
0.041 |
| 3 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
1.207 |
0.083 |
| 4 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
0.937 |
0.039 |
| 5 |
250624_at |
AT5G07330
|
unknown |
0.925 |
0.103 |
| 6 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
0.886 |
0.139 |
| 7 |
254823_at |
AT4G12580
|
unknown |
0.842 |
0.059 |
| 8 |
246241_at |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
0.727 |
0.059 |
| 9 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.719 |
0.109 |
| 10 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
0.664 |
-0.082 |
| 11 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.653 |
0.153 |
| 12 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.598 |
0.111 |
| 13 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.580 |
-0.019 |
| 14 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
0.577 |
0.085 |
| 15 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.551 |
-0.094 |
| 16 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.551 |
0.162 |
| 17 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.547 |
-0.299 |
| 18 |
261197_at |
AT1G12900
|
GAPA-2, glyceraldehyde 3-phosphate dehydrogenase A subunit 2 |
0.543 |
-0.230 |
| 19 |
257672_at |
AT3G20300
|
unknown |
0.539 |
-0.080 |
| 20 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.508 |
0.098 |
| 21 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.506 |
-0.498 |
| 22 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.498 |
0.125 |
| 23 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.472 |
-0.074 |
| 24 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.466 |
-0.301 |
| 25 |
260584_at |
AT2G43660
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.465 |
0.125 |
| 26 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
0.459 |
0.387 |
| 27 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.458 |
0.040 |
| 28 |
263881_at |
AT2G21820
|
unknown |
0.444 |
0.086 |
| 29 |
246617_at |
AT5G36270
|
[similar to DHAR2, glutathione dehydrogenase (ascorbate) [Arabidopsis thaliana] (TAIR:AT1G75270.1)] |
0.440 |
0.016 |
| 30 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.414 |
-0.199 |
| 31 |
245044_at |
AT2G26500
|
[cytochrome b6f complex subunit (petM), putative] |
0.404 |
-0.081 |
| 32 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.401 |
0.164 |
| 33 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
0.397 |
-0.360 |
| 34 |
252888_at |
AT4G39210
|
APL3 |
0.394 |
-0.110 |
| 35 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.387 |
0.184 |
| 36 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.387 |
0.268 |
| 37 |
267036_at |
AT2G38465
|
unknown |
0.377 |
-0.090 |
| 38 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.373 |
-0.061 |
| 39 |
256114_at |
AT1G16850
|
unknown |
0.372 |
0.156 |
| 40 |
250859_at |
AT5G04660
|
CYP77A4, cytochrome P450, family 77, subfamily A, polypeptide 4 |
0.365 |
0.051 |
| 41 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
0.359 |
0.123 |
| 42 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.357 |
-0.093 |
| 43 |
263709_at |
AT1G09310
|
unknown |
0.353 |
-0.678 |
| 44 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.351 |
-0.011 |
| 45 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.350 |
-0.030 |
| 46 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.348 |
-0.489 |
| 47 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.347 |
-0.247 |
| 48 |
247061_at |
AT5G66780
|
unknown |
0.347 |
0.098 |
| 49 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
0.342 |
-0.114 |
| 50 |
250611_at |
AT5G07200
|
ATGA20OX3, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 3, GA20OX3, gibberellin 20-oxidase 3, YAP169 |
0.341 |
0.101 |
| 51 |
253494_at |
AT4G31830
|
unknown |
0.336 |
0.018 |
| 52 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
0.333 |
0.074 |
| 53 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.848 |
-0.395 |
| 54 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-0.722 |
0.068 |
| 55 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.716 |
-0.042 |
| 56 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.635 |
-0.236 |
| 57 |
267137_at |
AT2G23410
|
ACPT, cis-prenyltransferase, AtcPT3, CPT, cis-prenyltransferase, cPT3, cis-prenyltransferase 3 |
-0.582 |
0.008 |
| 58 |
253044_at |
AT4G37290
|
unknown |
-0.566 |
0.262 |
| 59 |
245076_at |
AT2G23170
|
GH3.3 |
-0.553 |
0.484 |
| 60 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.547 |
0.007 |
| 61 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.546 |
-0.443 |
| 62 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.542 |
0.084 |
| 63 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.498 |
-0.070 |
| 64 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.464 |
-0.372 |
| 65 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.455 |
0.267 |
| 66 |
267085_at |
AT2G32620
|
ATCSLB02, cellulose synthase-like B, ATCSLB2, CELLULOSE SYNTHASE LIKE B2, CSLB02, CELLULOSE SYNTHASE LIKE B2, CSLB02, cellulose synthase-like B |
-0.452 |
0.069 |
| 67 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.444 |
0.036 |
| 68 |
262162_at |
AT1G78020
|
unknown |
-0.428 |
-0.350 |
| 69 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.410 |
0.233 |
| 70 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.395 |
-0.208 |
| 71 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.379 |
0.239 |
| 72 |
262091_at |
AT1G56160
|
ATMYB72, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 72, MYB72, myb domain protein 72 |
-0.372 |
0.100 |
| 73 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.370 |
0.207 |
| 74 |
262863_at |
AT1G64910
|
[UDP-Glycosyltransferase superfamily protein] |
-0.360 |
0.083 |
| 75 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.353 |
0.445 |
| 76 |
263515_at |
AT2G21640
|
unknown |
-0.342 |
0.077 |
| 77 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.326 |
-0.262 |
| 78 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.311 |
-0.057 |
| 79 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.290 |
-0.022 |
| 80 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.274 |
0.130 |
| 81 |
251293_at |
AT3G61930
|
unknown |
-0.263 |
0.030 |
| 82 |
253305_at |
AT4G33666
|
unknown |
-0.250 |
0.027 |
| 83 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
-0.249 |
0.016 |
| 84 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
-0.248 |
-0.061 |
| 85 |
267483_at |
AT2G02810
|
ATUTR1, UDP-GALACTOSE TRANSPORTER 1, UTR1, UDP-galactose transporter 1 |
-0.247 |
0.131 |
| 86 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.233 |
-0.228 |
| 87 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.232 |
-0.356 |
| 88 |
262373_at |
AT1G73120
|
unknown |
-0.231 |
0.056 |
| 89 |
246844_at |
AT5G26660
|
ATMYB86, myb domain protein 86, MYB86, myb domain protein 86 |
-0.230 |
0.291 |
| 90 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.230 |
0.175 |
| 91 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.222 |
-0.259 |
| 92 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.222 |
-0.484 |
| 93 |
253729_at |
AT4G29360
|
[O-Glycosyl hydrolases family 17 protein] |
-0.221 |
0.062 |
| 94 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-0.217 |
0.366 |
| 95 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.215 |
0.139 |
| 96 |
264013_at |
AT2G21070
|
FIO1, FIONA1 |
-0.206 |
0.128 |
| 97 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
-0.205 |
0.090 |
| 98 |
263683_at |
AT1G26870
|
ANAC009, Arabidopsis NAC domain containing protein 9, FEZ, FEZ |
-0.203 |
0.051 |
| 99 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.202 |
-0.041 |
| 100 |
256359_at |
AT1G66460
|
[Protein kinase superfamily protein] |
-0.200 |
0.098 |
| 101 |
251182_at |
AT3G62600
|
ATERDJ3B, ERDJ3B |
-0.199 |
0.151 |
| 102 |
245293_at |
AT4G16660
|
[heat shock protein 70 (Hsp 70) family protein] |
-0.196 |
0.166 |
| 103 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.193 |
0.108 |
| 104 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
-0.192 |
-0.053 |