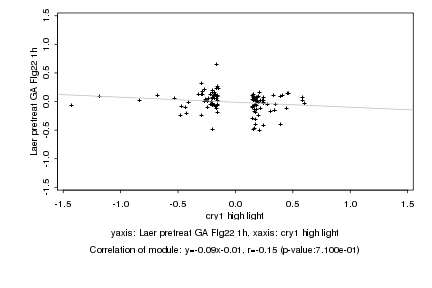
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cry1 high light |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.596 |
-0.022 |
| 2 |
262719_at |
AT1G43590
|
unknown |
0.579 |
0.021 |
| 3 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.578 |
0.084 |
| 4 |
253689_at |
AT4G29770
|
unknown |
0.455 |
0.145 |
| 5 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
0.449 |
0.156 |
| 6 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.439 |
-0.119 |
| 7 |
256418_at |
AT3G06160
|
[AP2/B3-like transcriptional factor family protein] |
0.408 |
0.117 |
| 8 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
0.385 |
-0.400 |
| 9 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.384 |
0.101 |
| 10 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.345 |
-0.040 |
| 11 |
247643_at |
AT5G60450
|
ARF4, auxin response factor 4 |
0.333 |
-0.148 |
| 12 |
250327_at |
AT5G12050
|
unknown |
0.332 |
-0.147 |
| 13 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.326 |
0.117 |
| 14 |
259373_at |
AT1G69160
|
unknown |
0.301 |
-0.169 |
| 15 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
0.278 |
-0.041 |
| 16 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
0.240 |
0.087 |
| 17 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
0.239 |
-0.411 |
| 18 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
0.238 |
-0.031 |
| 19 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.237 |
0.034 |
| 20 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
0.236 |
-0.007 |
| 21 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
0.227 |
0.021 |
| 22 |
247474_at |
AT5G62280
|
unknown |
0.218 |
0.026 |
| 23 |
267452_at |
AT2G33860
|
ARF3, AUXIN RESPONSE TRANSCRIPTION FACTOR 3, ETT, ETTIN |
0.210 |
-0.111 |
| 24 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
0.208 |
0.172 |
| 25 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
0.208 |
-0.498 |
| 26 |
247754_at |
AT5G59080
|
unknown |
0.198 |
-0.231 |
| 27 |
261502_at |
AT1G14440
|
AtHB31, homeobox protein 31, FTM2, FLORAL TRANSITION AT THE MERISTEM2, HB31, homeobox protein 31, ZHD4, ZINC FINGER HOMEODOMAIN 4 |
0.194 |
0.088 |
| 28 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.193 |
-0.003 |
| 29 |
250013_at |
AT5G18040
|
unknown |
0.192 |
0.013 |
| 30 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.187 |
0.074 |
| 31 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
0.182 |
-0.055 |
| 32 |
263002_at |
AT1G54200
|
unknown |
0.180 |
0.086 |
| 33 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
0.176 |
-0.138 |
| 34 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.176 |
0.004 |
| 35 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
0.173 |
0.043 |
| 36 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
0.170 |
-0.184 |
| 37 |
259689_x_at |
AT1G63130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.168 |
0.054 |
| 38 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
0.167 |
-0.397 |
| 39 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.166 |
-0.043 |
| 40 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
0.166 |
-0.309 |
| 41 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.163 |
-0.470 |
| 42 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
0.159 |
-0.128 |
| 43 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
0.159 |
0.067 |
| 44 |
246142_at |
AT5G19970
|
unknown |
0.158 |
-0.170 |
| 45 |
262399_at |
AT1G49500
|
unknown |
0.158 |
-0.067 |
| 46 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
0.156 |
-0.081 |
| 47 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
0.156 |
0.023 |
| 48 |
267580_at |
AT2G41990
|
unknown |
0.154 |
0.083 |
| 49 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
0.153 |
0.104 |
| 50 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.150 |
-0.488 |
| 51 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
0.149 |
0.130 |
| 52 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.147 |
-0.284 |
| 53 |
248167_at |
AT5G54530
|
unknown |
0.141 |
-0.071 |
| 54 |
259182_at |
AT3G01750
|
[Ankyrin repeat family protein] |
0.141 |
0.107 |
| 55 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
0.140 |
-0.089 |
| 56 |
267271_at |
AT2G02540
|
ATHB21, homeobox protein 21, HB21, homeobox protein 21, ZFHD4, ZINC FINGER HOMEODOMAIN 4, ZHD3, ZINC FINGER HOMEODOMAIN 3 |
0.140 |
0.052 |
| 57 |
256454_at |
AT1G75280
|
[NmrA-like negative transcriptional regulator family protein] |
-1.434 |
-0.063 |
| 58 |
256453_at |
AT1G75270
|
DHAR2, dehydroascorbate reductase 2 |
-1.190 |
0.088 |
| 59 |
255068_at |
AT4G08920
|
ATCRY1, BLU1, BLUE LIGHT UNINHIBITED 1, CRY1, cryptochrome 1, HY4, ELONGATED HYPOCOTYL 4, OOP2, OUT OF PHASE 2 |
-0.836 |
0.026 |
| 60 |
256452_at |
AT1G75240
|
AtHB33, homeobox protein 33, HB33, homeobox protein 33, ZHD5, zinc-finger homeodomain 5 |
-0.683 |
0.112 |
| 61 |
257595_at |
AT3G24750
|
unknown |
-0.539 |
0.059 |
| 62 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
-0.486 |
-0.241 |
| 63 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.471 |
-0.085 |
| 64 |
256509_at |
AT1G75300
|
[NmrA-like negative transcriptional regulator family protein] |
-0.438 |
-0.094 |
| 65 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.430 |
-0.205 |
| 66 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.410 |
-0.003 |
| 67 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.322 |
0.125 |
| 68 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.304 |
0.331 |
| 69 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-0.301 |
0.131 |
| 70 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.297 |
-0.243 |
| 71 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.295 |
0.132 |
| 72 |
265510_at |
AT2G05630
|
ATG8D |
-0.292 |
0.183 |
| 73 |
245560_at |
AT4G15480
|
UGT84A1 |
-0.278 |
0.211 |
| 74 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
-0.277 |
0.011 |
| 75 |
245126_at |
AT2G47460
|
ATMYB12, MYB DOMAIN PROTEIN 12, MYB12, myb domain protein 12, PFG1, PRODUCTION OF FLAVONOL GLYCOSIDES 1 |
-0.264 |
0.042 |
| 76 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
-0.260 |
0.021 |
| 77 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
-0.254 |
0.031 |
| 78 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
-0.250 |
0.008 |
| 79 |
267036_at |
AT2G38465
|
unknown |
-0.246 |
-0.090 |
| 80 |
249497_at |
AT5G39220
|
[alpha/beta-Hydrolases superfamily protein] |
-0.235 |
0.068 |
| 81 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.225 |
0.134 |
| 82 |
267070_at |
AT2G41000
|
[Chaperone DnaJ-domain superfamily protein] |
-0.218 |
-0.050 |
| 83 |
247641_at |
AT5G60540
|
ATPDX2, PYRIDOXINE BIOSYNTHESIS 2, EMB2407, EMBRYO DEFECTIVE 2407, PDX2, pyridoxine biosynthesis 2 |
-0.216 |
0.008 |
| 84 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.208 |
0.068 |
| 85 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.206 |
0.075 |
| 86 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
-0.205 |
-0.478 |
| 87 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.204 |
-0.031 |
| 88 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.202 |
-0.024 |
| 89 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.202 |
0.197 |
| 90 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
-0.201 |
-0.051 |
| 91 |
264287_at |
AT1G61930
|
unknown |
-0.201 |
-0.060 |
| 92 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.200 |
0.164 |
| 93 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
-0.195 |
0.116 |
| 94 |
256816_at |
AT3G21400
|
unknown |
-0.191 |
-0.053 |
| 95 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.189 |
0.057 |
| 96 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.188 |
0.166 |
| 97 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
-0.176 |
0.135 |
| 98 |
267066_at |
AT2G41040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.175 |
-0.087 |
| 99 |
259537_at |
AT1G12370
|
PHR1, photolyase 1, UVR2, UV RESISTANCE 2 |
-0.173 |
-0.111 |
| 100 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.171 |
0.660 |
| 101 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-0.171 |
0.234 |
| 102 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
-0.166 |
0.061 |
| 103 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
-0.166 |
0.105 |
| 104 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
-0.163 |
0.116 |
| 105 |
252010_at |
AT3G52740
|
unknown |
-0.163 |
-0.180 |
| 106 |
261514_at |
AT1G71870
|
[MATE efflux family protein] |
-0.162 |
0.095 |
| 107 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.162 |
-0.067 |
| 108 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.162 |
0.021 |
| 109 |
264637_at |
AT1G65560
|
[Zinc-binding dehydrogenase family protein] |
-0.162 |
-0.036 |
| 110 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
-0.161 |
0.279 |
| 111 |
251422_at |
AT3G60540
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
-0.156 |
0.231 |