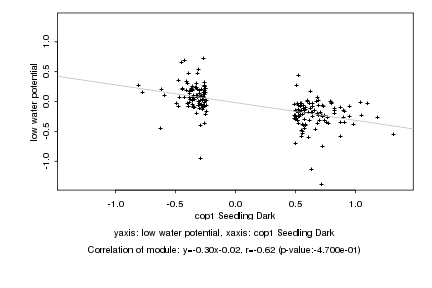
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cop1 Seedling Dark |
Log2 signal ratio
low water potential |
| 1 |
264636_at |
AT1G65490
|
unknown |
1.311 |
-0.540 |
| 2 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
1.178 |
-0.251 |
| 3 |
245025_at |
ATCG00130
|
ATPF |
1.094 |
-0.020 |
| 4 |
244934_at |
ATCG01080
|
NDHG |
1.045 |
-0.219 |
| 5 |
244933_at |
ATCG01070
|
NDHE |
1.041 |
-0.004 |
| 6 |
256796_at |
AT3G22210
|
unknown |
0.982 |
-0.372 |
| 7 |
244972_at |
ATCG00680
|
PSBB, photosystem II reaction center protein B |
0.948 |
-0.069 |
| 8 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
0.944 |
-0.245 |
| 9 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.907 |
-0.339 |
| 10 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.902 |
-0.160 |
| 11 |
244966_at |
ATCG00600
|
PETG |
0.893 |
-0.137 |
| 12 |
265611_at |
AT2G25510
|
unknown |
0.893 |
-0.265 |
| 13 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.874 |
-0.575 |
| 14 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.874 |
-0.341 |
| 15 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
0.870 |
-0.089 |
| 16 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
0.821 |
-0.139 |
| 17 |
263410_at |
AT2G04039
|
unknown |
0.819 |
-0.194 |
| 18 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
0.818 |
-0.111 |
| 19 |
245026_at |
ATCG00140
|
ATPH |
0.796 |
-0.014 |
| 20 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
0.793 |
-0.024 |
| 21 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
0.789 |
0.008 |
| 22 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
0.770 |
-0.365 |
| 23 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
0.764 |
-0.252 |
| 24 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
0.753 |
-0.338 |
| 25 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
0.737 |
-0.230 |
| 26 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
0.735 |
-0.317 |
| 27 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
0.726 |
-0.076 |
| 28 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
0.725 |
-0.061 |
| 29 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.724 |
-0.748 |
| 30 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
0.719 |
-0.236 |
| 31 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
0.714 |
-1.373 |
| 32 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
0.712 |
-0.231 |
| 33 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
0.704 |
-0.174 |
| 34 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
0.699 |
-0.313 |
| 35 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
0.692 |
-0.065 |
| 36 |
264201_at |
AT1G22630
|
unknown |
0.691 |
-0.234 |
| 37 |
260696_at |
AT1G32520
|
unknown |
0.687 |
0.023 |
| 38 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.680 |
-0.354 |
| 39 |
260004_at |
AT1G67860
|
unknown |
0.679 |
-0.189 |
| 40 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.679 |
0.072 |
| 41 |
245008_at |
ATCG00360
|
YCF3 |
0.670 |
0.015 |
| 42 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
0.660 |
-0.456 |
| 43 |
257966_at |
AT3G19800
|
unknown |
0.655 |
-0.146 |
| 44 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
0.641 |
-0.181 |
| 45 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.641 |
-0.073 |
| 46 |
247899_at |
AT5G57345
|
unknown |
0.638 |
-0.024 |
| 47 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
0.635 |
-0.241 |
| 48 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
0.634 |
-1.122 |
| 49 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.626 |
-0.172 |
| 50 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.621 |
0.181 |
| 51 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.619 |
-0.102 |
| 52 |
244998_at |
ATCG00180
|
RPOC1 |
0.616 |
-0.032 |
| 53 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
0.616 |
-0.305 |
| 54 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.609 |
-0.590 |
| 55 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.608 |
-0.177 |
| 56 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
0.595 |
0.003 |
| 57 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.594 |
0.024 |
| 58 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.581 |
-0.119 |
| 59 |
252876_at |
AT4G39970
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.581 |
-0.378 |
| 60 |
259625_at |
AT1G42970
|
GAPB, glyceraldehyde-3-phosphate dehydrogenase B subunit |
0.579 |
-0.096 |
| 61 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
0.576 |
-0.386 |
| 62 |
260368_at |
AT1G69700
|
ATHVA22C, HVA22 homologue C, HVA22C, HVA22 homologue C |
0.575 |
-0.168 |
| 63 |
260012_at |
AT1G67865
|
unknown |
0.573 |
-0.449 |
| 64 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
0.570 |
-0.143 |
| 65 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
0.570 |
-0.294 |
| 66 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.566 |
-0.398 |
| 67 |
245050_at |
ATCG00070
|
PSBK, photosystem II reaction center protein K precursor |
0.564 |
-0.107 |
| 68 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
0.561 |
-0.050 |
| 69 |
249932_at |
AT5G22390
|
unknown |
0.558 |
-0.487 |
| 70 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
0.557 |
-0.377 |
| 71 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.551 |
-0.203 |
| 72 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
0.551 |
-0.535 |
| 73 |
265569_at |
AT2G05620
|
AtPGR5, PGR5, proton gradient regulation 5 |
0.549 |
-0.150 |
| 74 |
252946_at |
AT4G39235
|
unknown |
0.547 |
-0.019 |
| 75 |
248623_at |
AT5G49170
|
unknown |
0.544 |
-0.478 |
| 76 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
0.543 |
-0.574 |
| 77 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.540 |
-0.082 |
| 78 |
245006_at |
ATCG00340
|
PSAB |
0.540 |
-0.026 |
| 79 |
262970_at |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
0.537 |
-0.212 |
| 80 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
0.537 |
-0.053 |
| 81 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
0.534 |
-0.242 |
| 82 |
260968_at |
AT1G12250
|
[Pentapeptide repeat-containing protein] |
0.528 |
-0.161 |
| 83 |
251031_at |
AT5G02120
|
OHP, one helix protein, PDE335, PIGMENT DEFECTIVE 335 |
0.524 |
-0.247 |
| 84 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
0.524 |
-0.058 |
| 85 |
250563_at |
AT5G08050
|
unknown |
0.523 |
-0.177 |
| 86 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.522 |
0.441 |
| 87 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.520 |
-0.130 |
| 88 |
250073_at |
AT5G17170
|
ENH1, enhancer of sos3-1 |
0.518 |
-0.358 |
| 89 |
260877_at |
AT1G21500
|
unknown |
0.514 |
-0.303 |
| 90 |
260542_at |
AT2G43560
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.514 |
-0.302 |
| 91 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
0.511 |
-0.021 |
| 92 |
251885_at |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
0.506 |
-0.162 |
| 93 |
259460_at |
AT1G44000
|
unknown |
0.505 |
-0.283 |
| 94 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.503 |
0.272 |
| 95 |
251784_at |
AT3G55330
|
PPL1, PsbP-like protein 1 |
0.500 |
-0.291 |
| 96 |
253790_at |
AT4G28660
|
PSB28, photosystem II reaction center PSB28 protein |
0.499 |
-0.229 |
| 97 |
254505_at |
AT4G19985
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.498 |
-0.247 |
| 98 |
258087_at |
AT3G26060
|
ATPRX Q, PRXQ, peroxiredoxin Q |
0.496 |
-0.136 |
| 99 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
0.495 |
-0.688 |
| 100 |
260704_at |
AT1G32470
|
[Single hybrid motif superfamily protein] |
0.492 |
-0.232 |
| 101 |
247853_at |
AT5G58140
|
NPL1, NON PHOTOTROPIC HYPOCOTYL 1-LIKE, PHOT2, phototropin 2 |
0.491 |
-0.038 |
| 102 |
251157_at |
AT3G63140
|
CSP41A, chloroplast stem-loop binding protein of 41 kDa |
0.491 |
-0.268 |
| 103 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
-0.810 |
0.277 |
| 104 |
259653_at |
AT1G55240
|
unknown |
-0.780 |
0.153 |
| 105 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.627 |
-0.435 |
| 106 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.619 |
0.207 |
| 107 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
-0.594 |
0.113 |
| 108 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.496 |
-0.028 |
| 109 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
-0.477 |
-0.077 |
| 110 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
-0.476 |
0.353 |
| 111 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.473 |
0.079 |
| 112 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
-0.457 |
0.658 |
| 113 |
248254_at |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
-0.447 |
0.204 |
| 114 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.446 |
0.225 |
| 115 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
-0.431 |
0.076 |
| 116 |
259897_at |
AT1G71380
|
ATCEL3, cellulase 3, ATGH9B3, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3, CEL3, cellulase 3 |
-0.429 |
0.688 |
| 117 |
255150_at |
AT4G08160
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
-0.413 |
0.196 |
| 118 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.411 |
0.345 |
| 119 |
246093_at |
AT5G20550
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.403 |
0.312 |
| 120 |
267311_at |
AT2G34670
|
unknown |
-0.402 |
-0.017 |
| 121 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.398 |
-0.082 |
| 122 |
246536_at |
AT5G15920
|
EMB2782, EMBRYO DEFECTIVE 2782, SMC5, structural maintenance of chromosomes 5 |
-0.397 |
-0.076 |
| 123 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
-0.392 |
0.480 |
| 124 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
-0.390 |
-0.009 |
| 125 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.389 |
0.079 |
| 126 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
-0.387 |
0.175 |
| 127 |
262795_at |
AT1G13130
|
[Cellulase (glycosyl hydrolase family 5) protein] |
-0.386 |
0.142 |
| 128 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
-0.375 |
0.037 |
| 129 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.374 |
0.220 |
| 130 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
-0.374 |
0.193 |
| 131 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.370 |
0.204 |
| 132 |
254632_at |
AT4G18630
|
unknown |
-0.365 |
0.252 |
| 133 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.365 |
0.211 |
| 134 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
-0.362 |
0.195 |
| 135 |
251478_at |
AT3G59690
|
IQD13, IQ-domain 13 |
-0.361 |
0.020 |
| 136 |
250756_at |
AT5G05940
|
ATROPGEF5, ROP GUANINE NUCLEOTIDE EXCHANGE FACTOR 5, ROPGEF5, ROP (rho of plants) guanine nucleotide exchange factor 5 |
-0.360 |
0.082 |
| 137 |
262873_at |
AT1G64700
|
unknown |
-0.359 |
-0.043 |
| 138 |
261031_at |
AT1G17360
|
unknown |
-0.356 |
-0.089 |
| 139 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
-0.355 |
0.109 |
| 140 |
253977_at |
AT4G26630
|
[DEK domain-containing chromatin associated protein] |
-0.355 |
0.040 |
| 141 |
266688_at |
AT2G19660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.351 |
-0.077 |
| 142 |
248150_at |
AT5G54670
|
ATK3, kinesin 3, KATC, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA C |
-0.348 |
0.040 |
| 143 |
245864_at |
AT1G58070
|
unknown |
-0.345 |
-0.085 |
| 144 |
263629_at |
AT2G04850
|
[Auxin-responsive family protein] |
-0.334 |
0.239 |
| 145 |
261491_at |
AT1G14350
|
AtMYB124, myb domain protein 124, FLP, FOUR LIPS, MYB124 |
-0.332 |
-0.196 |
| 146 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
-0.330 |
0.251 |
| 147 |
261500_at |
AT1G28400
|
unknown |
-0.330 |
0.115 |
| 148 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
-0.327 |
0.316 |
| 149 |
252319_at |
AT3G48710
|
[DEK domain-containing chromatin associated protein] |
-0.325 |
0.107 |
| 150 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
-0.323 |
0.210 |
| 151 |
248731_at |
AT5G48060
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.323 |
0.112 |
| 152 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.322 |
0.479 |
| 153 |
261879_at |
AT1G50520
|
CYP705A27, cytochrome P450, family 705, subfamily A, polypeptide 27 |
-0.319 |
0.074 |
| 154 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.319 |
0.074 |
| 155 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
-0.319 |
0.075 |
| 156 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.315 |
0.550 |
| 157 |
248070_at |
AT5G55660
|
[DEK domain-containing chromatin associated protein] |
-0.315 |
0.016 |
| 158 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
-0.314 |
-0.061 |
| 159 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
-0.313 |
-0.050 |
| 160 |
257534_at |
AT3G09670
|
[Tudor/PWWP/MBT superfamily protein] |
-0.307 |
-0.103 |
| 161 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
-0.304 |
-0.030 |
| 162 |
265083_at |
AT1G03820
|
unknown |
-0.302 |
0.201 |
| 163 |
255614_at |
AT4G01280
|
[Homeodomain-like superfamily protein] |
-0.300 |
0.143 |
| 164 |
249378_at |
AT5G40450
|
unknown |
-0.295 |
-0.399 |
| 165 |
249308_at |
AT5G41350
|
[RING/U-box superfamily protein] |
-0.295 |
0.025 |
| 166 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.294 |
-0.953 |
| 167 |
251967_at |
AT3G53390
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.293 |
0.087 |
| 168 |
253500_at |
AT4G31920
|
ARR10, response regulator 10, RR10, response regulator 10 |
-0.291 |
0.085 |
| 169 |
249939_at |
AT5G22430
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.290 |
-0.041 |
| 170 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
-0.287 |
0.216 |
| 171 |
264941_at |
AT1G60680
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.286 |
0.092 |
| 172 |
267488_at |
AT2G19110
|
ATHMA4, ARABIDOPSIS HEAVY METAL ATPASE 4, HMA4, heavy metal atpase 4 |
-0.284 |
-0.057 |
| 173 |
259367_at |
AT1G69070
|
unknown |
-0.283 |
0.030 |
| 174 |
258836_at |
AT3G07210
|
unknown |
-0.282 |
-0.118 |
| 175 |
246995_at |
AT5G67470
|
ATFH6, ARABIDOPSIS FORMIN HOMOLOG 6, FH6, formin homolog 6 |
-0.280 |
0.046 |
| 176 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
-0.279 |
-0.096 |
| 177 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.275 |
0.072 |
| 178 |
246826_at |
AT5G26310
|
UGT72E3 |
-0.274 |
0.044 |
| 179 |
250820_at |
AT5G05160
|
RUL1, REDUCED IN LATERAL GROWTH1 |
-0.273 |
0.092 |
| 180 |
251061_at |
AT5G01830
|
SAUR21, SMALL AUXIN UP RNA 21 |
-0.272 |
0.076 |
| 181 |
263253_at |
AT2G31370
|
[Basic-leucine zipper (bZIP) transcription factor family protein] |
-0.271 |
-0.058 |
| 182 |
261436_at |
AT1G07870
|
[Protein kinase superfamily protein] |
-0.271 |
0.132 |
| 183 |
267467_at |
AT2G30580
|
BMI1A, DRIP2, DREB2A-interacting protein 2 |
-0.270 |
-0.099 |
| 184 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.269 |
0.156 |
| 185 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.268 |
0.728 |
| 186 |
257217_at |
AT3G14940
|
ATPPC3, phosphoenolpyruvate carboxylase 3, PPC3, phosphoenolpyruvate carboxylase 3 |
-0.266 |
0.273 |
| 187 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
-0.265 |
0.262 |
| 188 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.265 |
0.122 |
| 189 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
-0.264 |
-0.356 |
| 190 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
-0.264 |
0.325 |
| 191 |
247312_at |
AT5G63970
|
RGLG3, RING DOMAIN LIGASE 3 |
-0.263 |
0.188 |
| 192 |
258828_at |
AT3G07130
|
ATPAP15, PURPLE ACID PHOSPHATASE 15, PAP15, purple acid phosphatase 15 |
-0.260 |
0.262 |
| 193 |
252667_at |
AT3G44200
|
ATNEK6, NIMA-RELATED KINASE6, IBO1, NEK6, NIMA (never in mitosis, gene A)-related 6 |
-0.260 |
-0.055 |
| 194 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.259 |
-0.005 |
| 195 |
251477_at |
AT3G59680
|
unknown |
-0.258 |
-0.082 |
| 196 |
252770_at |
AT3G42860
|
[zinc knuckle (CCHC-type) family protein] |
-0.257 |
0.167 |
| 197 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
-0.256 |
0.220 |
| 198 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
-0.255 |
0.233 |
| 199 |
266795_at |
AT2G03070
|
MED8, mediator subunit 8 |
-0.253 |
0.024 |
| 200 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
-0.252 |
-0.209 |
| 201 |
248998_at |
AT5G45320
|
unknown |
-0.249 |
0.026 |
| 202 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.248 |
-0.162 |
| 203 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.245 |
-0.057 |