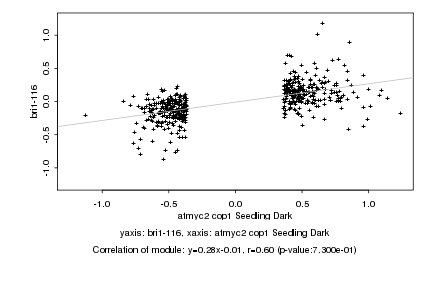
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
atmyc2 cop1 Seedling Dark |
Log2 signal ratio
bri1-116 |
| 1 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
1.236 |
-0.176 |
| 2 |
245025_at |
ATCG00130
|
ATPF |
1.138 |
0.055 |
| 3 |
264636_at |
AT1G65490
|
unknown |
1.095 |
0.172 |
| 4 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
1.080 |
0.094 |
| 5 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.011 |
-0.073 |
| 6 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
0.998 |
0.186 |
| 7 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.989 |
-0.257 |
| 8 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.959 |
-0.375 |
| 9 |
256796_at |
AT3G22210
|
unknown |
0.957 |
0.404 |
| 10 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.957 |
-0.082 |
| 11 |
265611_at |
AT2G25510
|
unknown |
0.909 |
0.067 |
| 12 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
0.882 |
0.151 |
| 13 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.870 |
0.254 |
| 14 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.854 |
0.890 |
| 15 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.846 |
-0.411 |
| 16 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.841 |
0.330 |
| 17 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
0.838 |
0.459 |
| 18 |
244933_at |
ATCG01070
|
NDHE |
0.834 |
0.040 |
| 19 |
260883_at |
AT1G29270
|
unknown |
0.818 |
0.553 |
| 20 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.811 |
0.098 |
| 21 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
0.791 |
0.066 |
| 22 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
0.790 |
-0.091 |
| 23 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.787 |
0.054 |
| 24 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.784 |
0.006 |
| 25 |
263410_at |
AT2G04039
|
unknown |
0.778 |
0.162 |
| 26 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
0.773 |
0.013 |
| 27 |
244934_at |
ATCG01080
|
NDHG |
0.771 |
0.018 |
| 28 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.769 |
0.647 |
| 29 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
0.767 |
0.047 |
| 30 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
0.763 |
0.101 |
| 31 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
0.758 |
0.149 |
| 32 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
0.753 |
0.256 |
| 33 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
0.753 |
0.273 |
| 34 |
244966_at |
ATCG00600
|
PETG |
0.752 |
0.010 |
| 35 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.740 |
0.136 |
| 36 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
0.737 |
0.256 |
| 37 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
0.726 |
0.631 |
| 38 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
0.720 |
0.123 |
| 39 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.717 |
0.169 |
| 40 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
0.713 |
0.025 |
| 41 |
245026_at |
ATCG00140
|
ATPH |
0.709 |
0.023 |
| 42 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.706 |
0.074 |
| 43 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
0.694 |
0.306 |
| 44 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
0.686 |
0.476 |
| 45 |
266293_at |
AT2G29360
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.671 |
0.300 |
| 46 |
261417_at |
AT1G07700
|
[Thioredoxin superfamily protein] |
0.669 |
0.186 |
| 47 |
245008_at |
ATCG00360
|
YCF3 |
0.668 |
0.042 |
| 48 |
260368_at |
AT1G69700
|
ATHVA22C, HVA22 homologue C, HVA22C, HVA22 homologue C |
0.668 |
0.124 |
| 49 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
0.666 |
-0.033 |
| 50 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.665 |
0.377 |
| 51 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.662 |
-0.268 |
| 52 |
253412_at |
AT4G33000
|
ATCBL10, CBL10, calcineurin B-like protein 10, SCABP8, SOS3-LIKE CALCIUM BINDING PROTEIN 8 |
0.661 |
0.285 |
| 53 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
0.651 |
0.220 |
| 54 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
0.651 |
0.291 |
| 55 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
0.649 |
1.183 |
| 56 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
0.639 |
0.282 |
| 57 |
244998_at |
ATCG00180
|
RPOC1 |
0.638 |
0.025 |
| 58 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
0.626 |
0.320 |
| 59 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
0.623 |
0.184 |
| 60 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
0.622 |
-0.062 |
| 61 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.619 |
0.027 |
| 62 |
252946_at |
AT4G39235
|
unknown |
0.611 |
0.208 |
| 63 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
0.609 |
1.019 |
| 64 |
251755_at |
AT3G55790
|
unknown |
0.608 |
-0.074 |
| 65 |
255440_at |
AT4G02530
|
[chloroplast thylakoid lumen protein] |
0.607 |
0.219 |
| 66 |
255793_at |
AT2G33250
|
unknown |
0.602 |
0.170 |
| 67 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
0.601 |
0.512 |
| 68 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
0.600 |
0.397 |
| 69 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.600 |
0.026 |
| 70 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
0.593 |
0.258 |
| 71 |
257191_at |
AT3G13175
|
unknown |
0.593 |
0.577 |
| 72 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.587 |
-0.022 |
| 73 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.586 |
0.121 |
| 74 |
261218_at |
AT1G20020
|
ATLFNR2, LEAF FNR 2, FNR2, ferredoxin-NADP(+)-oxidoreductase 2, LFNR2, leaf-type chloroplast-targeted FNR 2 |
0.580 |
0.080 |
| 75 |
248624_at |
AT5G48790
|
unknown |
0.580 |
0.263 |
| 76 |
259129_at |
AT3G02150
|
PTF1, plastid transcription factor 1, TCP13, TEOSINTE BRANCHED1, CYCLOIDEA AND PCF TRANSCRIPTION FACTOR 13, TFPD |
0.580 |
0.041 |
| 77 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
0.579 |
0.142 |
| 78 |
266658_at |
AT2G25735
|
unknown |
0.579 |
-0.232 |
| 79 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
0.578 |
-0.025 |
| 80 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.578 |
-0.014 |
| 81 |
260696_at |
AT1G32520
|
unknown |
0.578 |
0.095 |
| 82 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.573 |
0.157 |
| 83 |
257966_at |
AT3G19800
|
unknown |
0.572 |
0.216 |
| 84 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.564 |
0.218 |
| 85 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
0.563 |
0.081 |
| 86 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.561 |
0.322 |
| 87 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.559 |
0.332 |
| 88 |
260542_at |
AT2G43560
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.559 |
0.267 |
| 89 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
0.555 |
0.140 |
| 90 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
0.555 |
0.226 |
| 91 |
261951_at |
AT1G64490
|
[DEK, chromatin associated protein] |
0.549 |
-0.134 |
| 92 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.548 |
0.149 |
| 93 |
256389_at |
AT3G06220
|
[AP2/B3-like transcriptional factor family protein] |
0.546 |
0.213 |
| 94 |
245309_at |
AT4G15140
|
unknown |
0.544 |
0.007 |
| 95 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
0.540 |
-0.018 |
| 96 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
0.536 |
0.025 |
| 97 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
0.531 |
0.111 |
| 98 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
0.530 |
0.153 |
| 99 |
247899_at |
AT5G57345
|
unknown |
0.528 |
0.440 |
| 100 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
0.528 |
0.161 |
| 101 |
245019_at |
ATCG00530
|
YCF10 |
0.522 |
-0.029 |
| 102 |
250073_at |
AT5G17170
|
ENH1, enhancer of sos3-1 |
0.517 |
0.152 |
| 103 |
252876_at |
AT4G39970
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.516 |
0.095 |
| 104 |
251031_at |
AT5G02120
|
OHP, one helix protein, PDE335, PIGMENT DEFECTIVE 335 |
0.515 |
0.149 |
| 105 |
267220_at |
AT2G02500
|
ATMEPCT, ISPD, MCT, 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLTRANSFERASE |
0.513 |
0.146 |
| 106 |
244972_at |
ATCG00680
|
PSBB, photosystem II reaction center protein B |
0.510 |
0.114 |
| 107 |
255572_at |
AT4G01050
|
TROL, thylakoid rhodanese-like |
0.510 |
0.255 |
| 108 |
265569_at |
AT2G05620
|
AtPGR5, PGR5, proton gradient regulation 5 |
0.506 |
0.184 |
| 109 |
266916_at |
AT2G45860
|
unknown |
0.506 |
0.056 |
| 110 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.501 |
-0.347 |
| 111 |
251784_at |
AT3G55330
|
PPL1, PsbP-like protein 1 |
0.499 |
0.342 |
| 112 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
0.498 |
0.056 |
| 113 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.496 |
0.171 |
| 114 |
266074_at |
AT2G18740
|
[Small nuclear ribonucleoprotein family protein] |
0.494 |
-0.013 |
| 115 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
0.494 |
0.552 |
| 116 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.493 |
0.220 |
| 117 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
0.492 |
0.058 |
| 118 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.492 |
0.195 |
| 119 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.492 |
0.005 |
| 120 |
246270_at |
AT4G36500
|
unknown |
0.490 |
-0.225 |
| 121 |
251885_at |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
0.488 |
0.085 |
| 122 |
250563_at |
AT5G08050
|
unknown |
0.487 |
0.202 |
| 123 |
259625_at |
AT1G42970
|
GAPB, glyceraldehyde-3-phosphate dehydrogenase B subunit |
0.483 |
0.208 |
| 124 |
260704_at |
AT1G32470
|
[Single hybrid motif superfamily protein] |
0.482 |
0.180 |
| 125 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
0.481 |
0.233 |
| 126 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
0.481 |
0.181 |
| 127 |
266521_at |
AT2G24020
|
[Uncharacterised BCR, YbaB family COG0718] |
0.480 |
0.186 |
| 128 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
0.479 |
0.134 |
| 129 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.479 |
0.209 |
| 130 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.479 |
0.114 |
| 131 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
0.478 |
0.070 |
| 132 |
247853_at |
AT5G58140
|
NPL1, NON PHOTOTROPIC HYPOCOTYL 1-LIKE, PHOT2, phototropin 2 |
0.478 |
0.070 |
| 133 |
249253_at |
AT5G42060
|
[DEK, chromatin associated protein] |
0.478 |
-0.095 |
| 134 |
253635_at |
AT4G30620
|
[Uncharacterised BCR, YbaB family COG0718] |
0.476 |
0.280 |
| 135 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.475 |
0.004 |
| 136 |
244971_at |
ATCG00670
|
CLPP1, CASEINOLYTIC PROTEASE P 1, PCLPP, plastid-encoded CLP P |
0.474 |
0.019 |
| 137 |
248415_at |
AT5G51620
|
[Uncharacterised protein family (UPF0172)] |
0.472 |
0.071 |
| 138 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.472 |
0.088 |
| 139 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
0.472 |
0.234 |
| 140 |
249941_at |
AT5G22270
|
unknown |
0.472 |
0.011 |
| 141 |
245006_at |
ATCG00340
|
PSAB |
0.471 |
0.040 |
| 142 |
264635_at |
AT1G65500
|
unknown |
0.471 |
-0.190 |
| 143 |
253283_at |
AT4G34090
|
unknown |
0.470 |
0.378 |
| 144 |
263674_at |
AT2G04790
|
unknown |
0.468 |
0.302 |
| 145 |
263535_at |
AT2G24970
|
unknown |
0.468 |
-0.176 |
| 146 |
254043_at |
AT4G25990
|
CIL |
0.467 |
0.171 |
| 147 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.466 |
0.109 |
| 148 |
254505_at |
AT4G19985
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.463 |
0.154 |
| 149 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
0.462 |
0.138 |
| 150 |
256569_at |
AT3G19550
|
unknown |
0.459 |
0.058 |
| 151 |
262970_at |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
0.459 |
0.349 |
| 152 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.459 |
-0.178 |
| 153 |
262897_at |
AT1G59840
|
CCB4, cofactor assembly of complex C |
0.455 |
0.160 |
| 154 |
245050_at |
ATCG00070
|
PSBK, photosystem II reaction center protein K precursor |
0.454 |
0.013 |
| 155 |
260125_at |
AT1G36390
|
[Co-chaperone GrpE family protein] |
0.450 |
0.173 |
| 156 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
0.449 |
0.092 |
| 157 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.449 |
-0.132 |
| 158 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
0.448 |
0.443 |
| 159 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
0.446 |
-0.036 |
| 160 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.445 |
0.354 |
| 161 |
264054_at |
AT2G22540
|
AGL22, AGAMOUS-like 22, FAQ1, Flowering Arabidopsis QTL1, SVP, SHORT VEGETATIVE PHASE |
0.444 |
0.291 |
| 162 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.443 |
-0.116 |
| 163 |
265819_at |
AT2G17972
|
unknown |
0.443 |
0.195 |
| 164 |
256049_at |
AT1G07010
|
AtSLP1, SLP1, Shewenella-like protein phosphatase 1 |
0.442 |
0.122 |
| 165 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
0.441 |
0.381 |
| 166 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
0.440 |
-0.002 |
| 167 |
254642_at |
AT4G18810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.439 |
0.151 |
| 168 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
0.438 |
0.339 |
| 169 |
261039_at |
AT1G17455
|
ELF4-L4, ELF4-like 4 |
0.437 |
0.069 |
| 170 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
0.435 |
0.083 |
| 171 |
249875_at |
AT5G23120
|
HCF136, HIGH CHLOROPHYLL FLUORESCENCE 136 |
0.434 |
0.176 |
| 172 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.434 |
0.300 |
| 173 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.434 |
0.069 |
| 174 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
0.434 |
0.465 |
| 175 |
251980_at |
AT3G53270
|
[Small nuclear RNA activating complex (SNAPc), subunit SNAP43 protein] |
0.433 |
0.008 |
| 176 |
246905_at |
AT5G25570
|
unknown |
0.430 |
0.004 |
| 177 |
248277_at |
AT5G52860
|
ABCG8, ATP-binding cassette G8 |
0.429 |
0.155 |
| 178 |
245048_at |
ATCG00040
|
MATK, maturase K |
0.426 |
-0.001 |
| 179 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.426 |
0.248 |
| 180 |
248205_at |
AT5G54300
|
unknown |
0.424 |
0.095 |
| 181 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
0.424 |
0.047 |
| 182 |
266636_at |
AT2G35370
|
GDCH, glycine decarboxylase complex H |
0.422 |
0.135 |
| 183 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
0.422 |
0.177 |
| 184 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
0.422 |
0.046 |
| 185 |
260968_at |
AT1G12250
|
[Pentapeptide repeat-containing protein] |
0.421 |
0.254 |
| 186 |
267294_at |
AT2G23670
|
YCF37, homolog of Synechocystis YCF37 |
0.421 |
0.278 |
| 187 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.419 |
0.685 |
| 188 |
245004_at |
ATCG00300
|
YCF9 |
0.418 |
0.095 |
| 189 |
259132_at |
AT3G02250
|
[O-fucosyltransferase family protein] |
0.418 |
-0.025 |
| 190 |
249007_at |
AT5G44650
|
AtCEST, Arabidopsis thaliana chloroplast protein-enhancing stress tolerance, CEST, chloroplast protein-enhancing stress tolerance, Y3IP1, Ycf3-interacting protein 1 |
0.417 |
0.222 |
| 191 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
0.413 |
0.333 |
| 192 |
261611_at |
AT1G49730
|
[Protein kinase superfamily protein] |
0.413 |
-0.034 |
| 193 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.412 |
0.132 |
| 194 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
0.412 |
-0.103 |
| 195 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.411 |
0.166 |
| 196 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
0.411 |
0.429 |
| 197 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.408 |
-0.084 |
| 198 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.407 |
-0.042 |
| 199 |
246492_at |
AT5G16140
|
[Peptidyl-tRNA hydrolase family protein] |
0.406 |
0.254 |
| 200 |
259889_at |
AT1G76405
|
unknown |
0.406 |
0.326 |
| 201 |
254779_at |
AT4G12760
|
unknown |
0.405 |
0.106 |
| 202 |
258087_at |
AT3G26060
|
ATPRX Q, PRXQ, peroxiredoxin Q |
0.405 |
0.277 |
| 203 |
253917_at |
AT4G27380
|
unknown |
0.402 |
-0.028 |
| 204 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.402 |
-0.111 |
| 205 |
251810_at |
AT3G55250
|
PDE329, PIGMENT DEFECTIVE 329 |
0.402 |
0.196 |
| 206 |
259737_at |
AT1G64400
|
LACS3, long-chain acyl-CoA synthetase 3 |
0.401 |
0.013 |
| 207 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.401 |
0.702 |
| 208 |
251834_at |
AT3G55170
|
[Ribosomal L29 family protein ] |
0.399 |
0.159 |
| 209 |
263880_at |
AT2G21960
|
unknown |
0.397 |
0.171 |
| 210 |
252353_at |
AT3G48200
|
unknown |
0.393 |
0.157 |
| 211 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.391 |
0.303 |
| 212 |
249932_at |
AT5G22390
|
unknown |
0.388 |
0.141 |
| 213 |
261598_at |
AT1G49750
|
[Leucine-rich repeat (LRR) family protein] |
0.388 |
0.222 |
| 214 |
251701_at |
AT3G56650
|
PPD6, PsbP-domain protein 6 |
0.387 |
0.186 |
| 215 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.387 |
0.175 |
| 216 |
265967_at |
AT2G37450
|
UMAMIT13, Usually multiple acids move in and out Transporters 13 |
0.386 |
0.150 |
| 217 |
260877_at |
AT1G21500
|
unknown |
0.386 |
0.703 |
| 218 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
0.384 |
0.267 |
| 219 |
249482_at |
AT5G38980
|
unknown |
0.384 |
0.203 |
| 220 |
250128_at |
AT5G16540
|
ZFN3, zinc finger nuclease 3 |
0.384 |
0.113 |
| 221 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.384 |
0.186 |
| 222 |
256613_at |
AT3G29290
|
emb2076, embryo defective 2076 |
0.383 |
0.143 |
| 223 |
245326_at |
AT4G14100
|
[transferases, transferring glycosyl groups] |
0.382 |
0.144 |
| 224 |
261751_at |
AT1G76080
|
ATCDSP32, ARABIDOPSIS THALIANA CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD, CDSP32, chloroplastic drought-induced stress protein of 32 kD |
0.382 |
0.245 |
| 225 |
250531_at |
AT5G08650
|
[Small GTP-binding protein] |
0.381 |
0.085 |
| 226 |
267578_at |
AT2G30695
|
unknown |
0.381 |
0.164 |
| 227 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
0.381 |
0.320 |
| 228 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.381 |
0.057 |
| 229 |
253956_at |
AT4G26700
|
ATFIM1, ARABIDOPSIS THALIANA FIMBRIN 1, FIM1, fimbrin 1 |
0.380 |
0.196 |
| 230 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
0.380 |
0.178 |
| 231 |
248962_at |
AT5G45680
|
ATFKBP13, FK506 BINDING PROTEIN 13, FKBP13, FK506-binding protein 13 |
0.376 |
0.263 |
| 232 |
249276_at |
AT5G41880
|
POLA3, POLA4 |
0.375 |
-0.125 |
| 233 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
0.373 |
0.167 |
| 234 |
247800_at |
AT5G58570
|
unknown |
0.373 |
0.577 |
| 235 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.372 |
-0.126 |
| 236 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
0.370 |
0.178 |
| 237 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.370 |
0.037 |
| 238 |
245020_at |
ATCG00540
|
PETA, photosynthetic electron transfer A |
0.370 |
0.003 |
| 239 |
253796_at |
AT4G28460
|
unknown |
0.369 |
-0.181 |
| 240 |
251353_at |
AT3G61080
|
[Protein kinase superfamily protein] |
0.368 |
0.123 |
| 241 |
260506_at |
AT1G47210
|
CYCA3;2, cyclin-dependent protein kinase 3;2 |
0.367 |
-0.173 |
| 242 |
251157_at |
AT3G63140
|
CSP41A, chloroplast stem-loop binding protein of 41 kDa |
0.366 |
0.227 |
| 243 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
0.366 |
-0.004 |
| 244 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
0.366 |
0.238 |
| 245 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
0.364 |
-0.238 |
| 246 |
260106_at |
AT1G35420
|
[alpha/beta-Hydrolases superfamily protein] |
0.363 |
0.256 |
| 247 |
262598_at |
AT1G15260
|
unknown |
0.362 |
0.319 |
| 248 |
267526_at |
AT2G30570
|
PSBW, photosystem II reaction center W |
0.361 |
0.188 |
| 249 |
250482_at |
AT5G10320
|
unknown |
0.361 |
0.187 |
| 250 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.361 |
-0.070 |
| 251 |
263474_at |
AT2G31725
|
unknown |
0.359 |
-0.098 |
| 252 |
255663_at |
AT4G00420
|
[Double-stranded RNA-binding domain (DsRBD)-containing protein] |
0.358 |
0.032 |
| 253 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-1.130 |
-0.196 |
| 254 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.845 |
0.012 |
| 255 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
-0.793 |
-0.054 |
| 256 |
252347_at |
AT3G48130
|
RSU1 |
-0.772 |
0.084 |
| 257 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.768 |
-0.628 |
| 258 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.765 |
-0.459 |
| 259 |
259897_at |
AT1G71380
|
ATCEL3, cellulase 3, ATGH9B3, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3, CEL3, cellulase 3 |
-0.747 |
-0.324 |
| 260 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.733 |
-0.694 |
| 261 |
250230_at |
AT5G13900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.733 |
-0.070 |
| 262 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
-0.718 |
-0.790 |
| 263 |
251023_at |
AT5G02170
|
[Transmembrane amino acid transporter family protein] |
-0.716 |
-0.570 |
| 264 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.706 |
-0.099 |
| 265 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.701 |
-0.091 |
| 266 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
-0.695 |
-0.378 |
| 267 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.688 |
-0.403 |
| 268 |
250348_at |
AT5G11990
|
[proline-rich family protein] |
-0.685 |
-0.153 |
| 269 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
-0.683 |
-0.097 |
| 270 |
253822_at |
AT4G28410
|
RSA1, Root System Architecture 1 |
-0.675 |
-0.060 |
| 271 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-0.674 |
0.032 |
| 272 |
266546_at |
AT2G35270
|
AHL21, AT-hook motif nuclear localized protein 21, GIK, GIANT KILLER |
-0.671 |
-0.280 |
| 273 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.663 |
0.117 |
| 274 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.657 |
-0.257 |
| 275 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
-0.655 |
0.043 |
| 276 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
-0.651 |
-0.031 |
| 277 |
262349_at |
AT2G48130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.650 |
-0.141 |
| 278 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
-0.646 |
-0.092 |
| 279 |
249518_at |
AT5G38610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.639 |
-0.224 |
| 280 |
246913_at |
AT5G25830
|
GATA12, GATA transcription factor 12 |
-0.635 |
-0.059 |
| 281 |
253400_at |
AT4G32860
|
unknown |
-0.635 |
-0.133 |
| 282 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
-0.631 |
-0.281 |
| 283 |
267418_at |
AT2G35000
|
ATL9, Arabidopsis toxicos en levadura 9 |
-0.631 |
-0.161 |
| 284 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
-0.630 |
-0.171 |
| 285 |
250717_at |
AT5G06200
|
CASP4, Casparian strip membrane domain protein 4 |
-0.625 |
-0.293 |
| 286 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.625 |
-0.598 |
| 287 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.623 |
-0.114 |
| 288 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
-0.621 |
0.036 |
| 289 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.619 |
-0.110 |
| 290 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.617 |
-0.236 |
| 291 |
260024_at |
AT1G30080
|
[Glycosyl hydrolase superfamily protein] |
-0.617 |
-0.416 |
| 292 |
245140_at |
AT2G45420
|
LBD18, LOB domain-containing protein 18 |
-0.610 |
-0.058 |
| 293 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.610 |
-0.209 |
| 294 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
-0.609 |
-0.028 |
| 295 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.600 |
-0.308 |
| 296 |
253556_at |
AT4G31100
|
[wall-associated kinase, putative] |
-0.596 |
-0.102 |
| 297 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.595 |
-0.019 |
| 298 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.588 |
-0.351 |
| 299 |
251857_at |
AT3G54770
|
ARP1, ABA-regulated RNA-binding protein 1 |
-0.588 |
-0.362 |
| 300 |
248750_at |
AT5G47530
|
[Auxin-responsive family protein] |
-0.585 |
-0.177 |
| 301 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.585 |
-0.293 |
| 302 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
-0.582 |
-0.024 |
| 303 |
263734_at |
AT1G60030
|
ATNAT7, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 7, NAT7, nucleobase-ascorbate transporter 7 |
-0.579 |
-0.316 |
| 304 |
255646_at |
AT4G00890
|
[proline-rich family protein] |
-0.579 |
0.063 |
| 305 |
246235_at |
AT4G36830
|
HOS3-1 |
-0.575 |
-0.168 |
| 306 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
-0.572 |
-0.152 |
| 307 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
-0.568 |
-0.143 |
| 308 |
262795_at |
AT1G13130
|
[Cellulase (glycosyl hydrolase family 5) protein] |
-0.568 |
-0.168 |
| 309 |
246476_at |
AT5G16730
|
[LOCATED IN: chloroplast] |
-0.567 |
-0.017 |
| 310 |
248621_at |
AT5G49350
|
[Glycine-rich protein family] |
-0.562 |
0.009 |
| 311 |
254206_at |
AT4G24180
|
ATTLP1, TLP1, THAUMATIN-LIKE PROTEIN 1 |
-0.561 |
-0.318 |
| 312 |
267343_at |
AT2G44260
|
unknown |
-0.558 |
-0.102 |
| 313 |
247354_at |
AT5G63590
|
ATFLS3, FLS3, flavonol synthase 3 |
-0.557 |
-0.368 |
| 314 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.556 |
0.190 |
| 315 |
266978_at |
AT2G39430
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.556 |
-0.328 |
| 316 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
-0.555 |
-0.069 |
| 317 |
265645_at |
AT2G27370
|
CASP3, Casparian strip membrane domain protein 3 |
-0.554 |
-0.308 |
| 318 |
258795_at |
AT3G04570
|
AHL19, AT-hook motif nuclear-localized protein 19 |
-0.552 |
-0.029 |
| 319 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.550 |
-0.224 |
| 320 |
250674_at |
AT5G07130
|
LAC13, laccase 13 |
-0.550 |
0.155 |
| 321 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.548 |
-0.153 |
| 322 |
261640_at |
AT1G49960
|
[Xanthine/uracil permease family protein] |
-0.545 |
0.017 |
| 323 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.545 |
-0.862 |
| 324 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
-0.544 |
-0.093 |
| 325 |
248254_at |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
-0.542 |
-0.010 |
| 326 |
258745_at |
AT3G05920
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.537 |
-0.208 |
| 327 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
-0.537 |
-0.407 |
| 328 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
-0.537 |
0.179 |
| 329 |
252400_at |
AT3G48020
|
unknown |
-0.536 |
-0.196 |
| 330 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.535 |
-0.162 |
| 331 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.535 |
-0.029 |
| 332 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.533 |
-0.149 |
| 333 |
253626_at |
AT4G30640
|
[RNI-like superfamily protein] |
-0.531 |
-0.102 |
| 334 |
259291_at |
AT3G11550
|
CASP2, Casparian strip membrane domain protein 2 |
-0.530 |
-0.299 |
| 335 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
-0.527 |
-0.037 |
| 336 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.526 |
-0.727 |
| 337 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.521 |
-0.062 |
| 338 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.519 |
-0.076 |
| 339 |
265131_at |
AT1G23760
|
JP630, PG3, POLYGALACTURONASE 3 |
-0.518 |
-0.233 |
| 340 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.517 |
0.039 |
| 341 |
256674_at |
AT3G52360
|
unknown |
-0.515 |
-0.238 |
| 342 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
-0.514 |
-0.136 |
| 343 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
-0.514 |
-0.131 |
| 344 |
248761_at |
AT5G47635
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.512 |
-0.038 |
| 345 |
261500_at |
AT1G28400
|
unknown |
-0.512 |
-0.237 |
| 346 |
249167_at |
AT5G42860
|
unknown |
-0.511 |
-0.338 |
| 347 |
246425_at |
AT5G17420
|
ATCESA7, CESA7, CELLULOSE SYNTHASE CATALYTIC SUBUNIT 7, IRX3, IRREGULAR XYLEM 3, MUR10, MURUS 10 |
-0.511 |
-0.109 |
| 348 |
249729_at |
AT5G24410
|
PGL4, 6-phosphogluconolactonase 4 |
-0.510 |
-0.013 |
| 349 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
-0.508 |
-0.032 |
| 350 |
251660_at |
AT3G57160
|
unknown |
-0.505 |
0.043 |
| 351 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
-0.504 |
-0.128 |
| 352 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.503 |
-0.402 |
| 353 |
266425_at |
AT2G41300
|
SSL1, strictosidine synthase-like 1 |
-0.501 |
-0.236 |
| 354 |
253864_at |
AT4G27460
|
CBSX5, CBS domain containing protein 5 |
-0.500 |
-0.118 |
| 355 |
266356_at |
AT2G32300
|
UCC1, uclacyanin 1 |
-0.498 |
-0.203 |
| 356 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.498 |
-0.303 |
| 357 |
256750_at |
AT3G27150
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.496 |
0.004 |
| 358 |
256217_at |
AT1G56320
|
unknown |
-0.492 |
-0.025 |
| 359 |
248070_at |
AT5G55660
|
[DEK domain-containing chromatin associated protein] |
-0.492 |
-0.091 |
| 360 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
-0.491 |
0.076 |
| 361 |
261878_at |
AT1G50560
|
CYP705A25, cytochrome P450, family 705, subfamily A, polypeptide 25 |
-0.491 |
-0.603 |
| 362 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.489 |
-0.049 |
| 363 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
-0.489 |
-0.168 |
| 364 |
245556_at |
AT4G15400
|
ABS1, ABNORMAL SHOOT 1, BIA1, BRASSINOSTEROID INACTIVATOR1 |
-0.487 |
-0.094 |
| 365 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.487 |
0.026 |
| 366 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
-0.487 |
0.048 |
| 367 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
-0.484 |
-0.474 |
| 368 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
-0.484 |
-0.132 |
| 369 |
249070_at |
AT5G44030
|
CESA4, cellulose synthase A4, IRX5, IRREGULAR XYLEM 5, NWS2 |
-0.483 |
-0.085 |
| 370 |
254632_at |
AT4G18630
|
unknown |
-0.481 |
-0.084 |
| 371 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.479 |
-0.108 |
| 372 |
246826_at |
AT5G26310
|
UGT72E3 |
-0.479 |
-0.322 |
| 373 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.479 |
-0.229 |
| 374 |
259268_at |
AT3G01070
|
AtENODL16, ENODL16, early nodulin-like protein 16 |
-0.478 |
-0.136 |
| 375 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.474 |
-0.023 |
| 376 |
261844_at |
AT1G15940
|
[Tudor/PWWP/MBT superfamily protein] |
-0.473 |
0.016 |
| 377 |
262662_at |
AT1G13920
|
[Remorin family protein] |
-0.473 |
0.045 |
| 378 |
247958_at |
AT5G57070
|
[hydroxyproline-rich glycoprotein family protein] |
-0.469 |
-0.243 |
| 379 |
251093_at |
AT5G01360
|
TBL3, TRICHOME BIREFRINGENCE-LIKE 3 |
-0.468 |
-0.197 |
| 380 |
261773_at |
AT1G76250
|
unknown |
-0.467 |
-0.156 |
| 381 |
245737_at |
AT1G44160
|
[HSP40/DnaJ peptide-binding protein] |
-0.466 |
-0.252 |
| 382 |
261879_at |
AT1G50520
|
CYP705A27, cytochrome P450, family 705, subfamily A, polypeptide 27 |
-0.466 |
-0.069 |
| 383 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.466 |
-0.244 |
| 384 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.466 |
-0.361 |
| 385 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
-0.465 |
-0.221 |
| 386 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
-0.464 |
-0.075 |
| 387 |
258878_at |
AT3G03170
|
unknown |
-0.462 |
-0.022 |
| 388 |
260484_at |
AT1G68360
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.461 |
-0.230 |
| 389 |
246387_at |
AT1G77400
|
unknown |
-0.460 |
0.007 |
| 390 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.457 |
0.101 |
| 391 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
-0.456 |
-0.288 |
| 392 |
249939_at |
AT5G22430
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.454 |
0.036 |
| 393 |
249378_at |
AT5G40450
|
unknown |
-0.453 |
-0.140 |
| 394 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.453 |
0.101 |
| 395 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
-0.452 |
-0.197 |
| 396 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.451 |
-0.768 |
| 397 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
-0.451 |
-0.024 |
| 398 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
-0.449 |
0.112 |
| 399 |
253985_at |
AT4G26220
|
CCoAOMT7, caffeoyl coenzyme A ester O-methyltransferase 7 |
-0.449 |
-0.228 |
| 400 |
257217_at |
AT3G14940
|
ATPPC3, phosphoenolpyruvate carboxylase 3, PPC3, phosphoenolpyruvate carboxylase 3 |
-0.449 |
-0.017 |
| 401 |
256447_at |
AT1G33440
|
AtNPF4.4, NPF4.4, NRT1/ PTR family 4.4 |
-0.448 |
0.202 |
| 402 |
263284_at |
AT2G36100
|
CASP1, Casparian strip membrane domain protein 1 |
-0.447 |
-0.192 |
| 403 |
253977_at |
AT4G26630
|
[DEK domain-containing chromatin associated protein] |
-0.447 |
-0.027 |
| 404 |
260163_at |
AT1G79900
|
ATMBAC2, RABIDOPSIS MITOCHONDRIAL BASIC AMINO ACID CARRIER 2, BAC2 |
-0.446 |
0.008 |
| 405 |
263112_at |
AT1G03080
|
NET1D, Networked 1D |
-0.445 |
-0.003 |
| 406 |
255873_at |
AT2G30340
|
LBD13, LOB domain-containing protein 13 |
-0.444 |
0.004 |
| 407 |
255528_at |
AT4G02090
|
unknown |
-0.443 |
-0.119 |
| 408 |
258828_at |
AT3G07130
|
ATPAP15, PURPLE ACID PHOSPHATASE 15, PAP15, purple acid phosphatase 15 |
-0.442 |
0.107 |
| 409 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
-0.441 |
-0.467 |
| 410 |
248121_at |
AT5G54690
|
GAUT12, galacturonosyltransferase 12, IRX8, IRREGULAR XYLEM 8, LGT6 |
-0.439 |
-0.148 |
| 411 |
253713_at |
AT4G29370
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.439 |
0.233 |
| 412 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.436 |
-0.733 |
| 413 |
259424_at |
AT1G13830
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.436 |
-0.249 |
| 414 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.435 |
-0.141 |
| 415 |
252458_at |
AT3G47210
|
unknown |
-0.433 |
-0.188 |
| 416 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
-0.432 |
-0.242 |
| 417 |
250756_at |
AT5G05940
|
ATROPGEF5, ROP GUANINE NUCLEOTIDE EXCHANGE FACTOR 5, ROPGEF5, ROP (rho of plants) guanine nucleotide exchange factor 5 |
-0.431 |
-0.099 |
| 418 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.430 |
-0.168 |
| 419 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
-0.429 |
-0.142 |
| 420 |
253100_at |
AT4G37400
|
CYP81F3, cytochrome P450, family 81, subfamily F, polypeptide 3 |
-0.429 |
-0.169 |
| 421 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-0.429 |
-0.152 |
| 422 |
245305_at |
AT4G17215
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.428 |
-0.039 |
| 423 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
-0.428 |
-0.168 |
| 424 |
246933_at |
AT5G25160
|
ZFP3, zinc finger protein 3 |
-0.427 |
-0.262 |
| 425 |
250438_at |
AT5G10580
|
unknown |
-0.426 |
-0.052 |
| 426 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.426 |
-0.176 |
| 427 |
265014_at |
AT1G24430
|
[HXXXD-type acyl-transferase family protein] |
-0.425 |
0.043 |
| 428 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.425 |
-0.531 |
| 429 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.425 |
-0.116 |
| 430 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
-0.424 |
-0.243 |
| 431 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
-0.423 |
-0.423 |
| 432 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
-0.422 |
-0.209 |
| 433 |
245788_at |
AT1G32120
|
unknown |
-0.422 |
-0.000 |
| 434 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
-0.421 |
-0.179 |
| 435 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
-0.420 |
-0.168 |
| 436 |
249886_at |
AT5G22320
|
[Leucine-rich repeat (LRR) family protein] |
-0.420 |
0.066 |
| 437 |
251478_at |
AT3G59690
|
IQD13, IQ-domain 13 |
-0.420 |
-0.368 |
| 438 |
253112_at |
AT4G35970
|
APX5, ascorbate peroxidase 5 |
-0.417 |
-0.221 |
| 439 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.414 |
0.091 |
| 440 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
-0.414 |
0.005 |
| 441 |
261673_at |
AT1G18280
|
LTPG3, glycosylphosphatidylinositol-anchored lipid protein transfer 3 |
-0.413 |
0.026 |
| 442 |
263937_at |
AT2G35910
|
[RING/U-box superfamily protein] |
-0.413 |
0.088 |
| 443 |
256362_at |
AT1G66450
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.412 |
-0.187 |
| 444 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.412 |
-0.204 |
| 445 |
251061_at |
AT5G01830
|
SAUR21, SMALL AUXIN UP RNA 21 |
-0.412 |
-0.249 |
| 446 |
248731_at |
AT5G48060
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.411 |
-0.033 |
| 447 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
-0.410 |
-0.103 |
| 448 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.408 |
-0.381 |
| 449 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
-0.405 |
-0.122 |
| 450 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.405 |
-0.226 |
| 451 |
246888_at |
AT5G26270
|
unknown |
-0.402 |
-0.231 |
| 452 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.400 |
-0.231 |
| 453 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.400 |
-0.529 |
| 454 |
261545_at |
AT1G63530
|
unknown |
-0.400 |
-0.261 |
| 455 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
-0.400 |
-0.099 |
| 456 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.398 |
-0.294 |
| 457 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
-0.398 |
-0.239 |
| 458 |
260666_at |
AT1G19300
|
ATGATL1, GATL1, GALACTURONOSYLTRANSFERASE-LIKE 1, GLZ1, GAOLAOZHUANGREN 1, PARVUS, PARVUS |
-0.396 |
-0.121 |
| 459 |
253424_at |
AT4G32330
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.396 |
-0.043 |
| 460 |
247394_at |
AT5G62865
|
unknown |
-0.396 |
-0.073 |
| 461 |
250070_at |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.395 |
-0.203 |
| 462 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
-0.395 |
0.058 |
| 463 |
259661_at |
AT1G55265
|
unknown |
-0.391 |
-0.250 |
| 464 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
-0.391 |
-0.261 |
| 465 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
-0.391 |
-0.279 |
| 466 |
249289_at |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
-0.390 |
-0.155 |
| 467 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.389 |
-0.108 |
| 468 |
257975_at |
AT3G20830
|
AGC2-4, AGC2 kinase 4, UCNL, UNICORN-like |
-0.388 |
-0.073 |
| 469 |
262318_at |
AT1G27620
|
[HXXXD-type acyl-transferase family protein] |
-0.388 |
-0.145 |
| 470 |
267311_at |
AT2G34670
|
unknown |
-0.388 |
-0.133 |
| 471 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.387 |
-0.090 |
| 472 |
256435_at |
AT3G11180
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.387 |
-0.055 |
| 473 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.386 |
-0.241 |
| 474 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.385 |
-0.148 |
| 475 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
-0.385 |
0.081 |
| 476 |
249790_at |
AT5G24290
|
MEB2, MEMBRANE OF ER BODY 2 |
-0.385 |
-0.436 |
| 477 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.385 |
-0.233 |
| 478 |
244923_s_at |
ATMG01020
|
ORF153B |
-0.384 |
-0.027 |
| 479 |
249838_at |
AT5G23460
|
unknown |
-0.383 |
0.096 |
| 480 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
-0.383 |
-0.092 |
| 481 |
250065_at |
AT5G17910
|
unknown |
-0.383 |
-0.196 |
| 482 |
250689_at |
AT5G06610
|
unknown |
-0.381 |
-0.062 |
| 483 |
254618_at |
AT4G18780
|
ATCESA8, CELLULOSE SYNTHASE 8, CESA8, CELLULOSE SYNTHASE 8, IRX1, IRREGULAR XYLEM 1, LEW2, LEAF WILTING 2 |
-0.380 |
-0.099 |
| 484 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.380 |
-0.129 |
| 485 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
-0.380 |
-0.303 |
| 486 |
247091_at |
AT5G66390
|
PRX72, PEROXIDASE 72 |
-0.379 |
-0.281 |
| 487 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
-0.378 |
0.052 |
| 488 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.378 |
-0.539 |
| 489 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.378 |
-0.043 |
| 490 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
-0.378 |
0.106 |
| 491 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
-0.376 |
-0.006 |
| 492 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.376 |
-0.119 |
| 493 |
253140_at |
AT4G35480
|
RHA3B, RING-H2 finger A3B |
-0.376 |
0.023 |
| 494 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
-0.376 |
-0.129 |
| 495 |
266581_at |
AT2G46140
|
[Late embryogenesis abundant protein] |
-0.375 |
-0.232 |
| 496 |
251078_at |
AT5G01990
|
[Auxin efflux carrier family protein] |
-0.374 |
-0.050 |
| 497 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
-0.374 |
-0.093 |
| 498 |
257147_at |
AT3G27270
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-0.373 |
-0.120 |
| 499 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.373 |
0.054 |
| 500 |
256025_at |
AT1G58370
|
ATXYN1, ARABIDOPSIS THALIANA XYLANASE 1, RXF12 |
-0.373 |
-0.138 |
| 501 |
245139_at |
AT2G45430
|
AHL22, AT-hook motif nuclear-localized protein 22 |
-0.373 |
-0.066 |
| 502 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.372 |
0.027 |
| 503 |
249958_at |
AT5G18970
|
[AWPM-19-like family protein] |
-0.372 |
-0.183 |