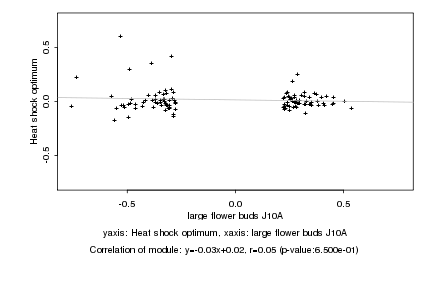
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
large flower buds J10A |
Log2 signal ratio
Heat shock optimum |
| 1 |
250619_at |
AT5G07230
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.535 |
-0.063 |
| 2 |
263029_at |
AT1G24050
|
[RNA-processing, Lsm domain] |
0.504 |
0.008 |
| 3 |
265883_at |
AT2G42310
|
unknown |
0.451 |
0.046 |
| 4 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.449 |
-0.014 |
| 5 |
266226_at |
AT2G28740
|
HIS4, histone H4 |
0.446 |
-0.026 |
| 6 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.420 |
0.055 |
| 7 |
259568_at |
AT1G20490
|
[AMP-dependent synthetase and ligase family protein] |
0.411 |
-0.036 |
| 8 |
245022_at |
ATCG00560
|
PSBL, photosystem II reaction center protein L |
0.405 |
-0.015 |
| 9 |
246351_at |
AT1G16570
|
[UDP-Glycosyltransferase superfamily protein] |
0.395 |
0.045 |
| 10 |
261244_at |
AT1G20150
|
[Subtilisin-like serine endopeptidase family protein] |
0.382 |
-0.029 |
| 11 |
247433_at |
AT5G62540
|
UBC3, ubiquitin-conjugating enzyme 3 |
0.375 |
0.005 |
| 12 |
262784_at |
AT1G10760
|
GWD, GWD1, SEX1, STARCH EXCESS 1, SOP, SOP1 |
0.372 |
0.070 |
| 13 |
262236_at |
AT1G48330
|
unknown |
0.365 |
0.076 |
| 14 |
251078_at |
AT5G01990
|
[Auxin efflux carrier family protein] |
0.350 |
-0.004 |
| 15 |
259986_at |
AT1G75050
|
[Pathogenesis-related thaumatin superfamily protein] |
0.348 |
-0.034 |
| 16 |
255545_at |
AT4G01890
|
[Pectin lyase-like superfamily protein] |
0.346 |
-0.021 |
| 17 |
246457_at |
AT5G16750
|
TOZ, TORMOZEMBRYO DEFECTIVE |
0.339 |
0.042 |
| 18 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
0.338 |
-0.026 |
| 19 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
0.328 |
0.001 |
| 20 |
247891_at |
AT5G57960
|
[GTP-binding protein, HflX] |
0.322 |
-0.110 |
| 21 |
252290_at |
AT3G49140
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.320 |
-0.021 |
| 22 |
266229_at |
AT2G28840
|
XBAT31, XB3 ortholog 1 in Arabidopsis thaliana |
0.317 |
0.053 |
| 23 |
263496_at |
AT2G42570
|
TBL39, TRICHOME BIREFRINGENCE-LIKE 39 |
0.317 |
0.088 |
| 24 |
258562_at |
AT3G05980
|
unknown |
0.316 |
-0.025 |
| 25 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
0.301 |
0.059 |
| 26 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.295 |
0.013 |
| 27 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
0.294 |
-0.016 |
| 28 |
250530_at |
AT5G08630
|
[DDT domain-containing protein] |
0.287 |
-0.017 |
| 29 |
252350_at |
AT3G48190
|
ATATM, ARABIDOPSIS THALIANA ATAXIA-TELANGIECTASIA MUTATED, ATM, ataxia-telangiectasia mutated, PIG1, pcd in male gametogenesis 1 |
0.286 |
-0.015 |
| 30 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.286 |
0.259 |
| 31 |
265898_at |
AT2G25690
|
unknown |
0.282 |
-0.050 |
| 32 |
258223_at |
AT3G15840
|
PIFI, post-illumination chlorophyll fluorescence increase |
0.279 |
0.016 |
| 33 |
254973_at |
AT4G10460
|
[copia-like retrotransposon family, has a 3.8e-291 P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.277 |
-0.009 |
| 34 |
248942_at |
AT5G45480
|
unknown |
0.276 |
-0.040 |
| 35 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
0.271 |
0.044 |
| 36 |
263348_at |
AT2G05710
|
ACO3, aconitase 3 |
0.270 |
-0.007 |
| 37 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
0.270 |
0.060 |
| 38 |
265916_at |
AT3G31920
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 3.0e-163 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.265 |
-0.055 |
| 39 |
260395_at |
AT1G69780
|
ATHB13 |
0.261 |
0.191 |
| 40 |
266180_at |
AT2G02470
|
AL6, alfin-like 6 |
0.260 |
0.009 |
| 41 |
248495_at |
AT5G50780
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
0.255 |
0.034 |
| 42 |
246436_at |
AT5G17440
|
[LUC7 related protein] |
0.254 |
0.023 |
| 43 |
267536_at |
AT2G42010
|
PLDBETA, PLDBETA1, phospholipase D beta 1 |
0.252 |
0.019 |
| 44 |
262397_at |
AT1G49380
|
[cytochrome c biogenesis protein family] |
0.249 |
-0.075 |
| 45 |
251864_at |
AT3G54920
|
PMR6, powdery mildew resistant 6 |
0.247 |
0.042 |
| 46 |
266727_at |
AT2G03150
|
emb1579, embryo defective 1579 |
0.246 |
-0.039 |
| 47 |
266796_at |
AT2G02880
|
[mucin-related] |
0.245 |
0.050 |
| 48 |
261757_at |
AT1G08210
|
[Eukaryotic aspartyl protease family protein] |
0.240 |
0.090 |
| 49 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
0.240 |
-0.009 |
| 50 |
249900_at |
AT5G22640
|
emb1211, embryo defective 1211, TIC100, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 100 |
0.236 |
-0.033 |
| 51 |
260242_at |
AT1G63650
|
AtEGL3, ATMYC-2, EGL1, EGL3, ENHANCER OF GLABRA 3 |
0.233 |
0.078 |
| 52 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
0.228 |
-0.056 |
| 53 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
0.226 |
0.045 |
| 54 |
255008_at |
AT4G10060
|
[Beta-glucosidase, GBA2 type family protein] |
0.226 |
-0.041 |
| 55 |
257717_at |
AT3G18390
|
EMB1865, embryo defective 1865 |
0.226 |
-0.023 |
| 56 |
261353_at |
AT1G79600
|
ABC1K3, ABC1-like kinase 3 |
0.224 |
-0.068 |
| 57 |
253322_at |
AT4G33980
|
unknown |
0.222 |
-0.047 |
| 58 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.221 |
0.035 |
| 59 |
252606_at |
AT3G45010
|
scpl48, serine carboxypeptidase-like 48 |
-0.761 |
-0.038 |
| 60 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
-0.737 |
0.232 |
| 61 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
-0.576 |
0.054 |
| 62 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.562 |
-0.172 |
| 63 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
-0.552 |
-0.061 |
| 64 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.534 |
0.610 |
| 65 |
250126_at |
AT5G16310
|
UCH1 |
-0.527 |
-0.034 |
| 66 |
263426_at |
AT2G31570
|
ATGPX2, glutathione peroxidase 2, GPX2, glutathione peroxidase 2 |
-0.518 |
-0.036 |
| 67 |
262645_at |
AT1G62750
|
ATSCO1, SNOWY COTYLEDON 1, ATSCO1/CPEF-G, SCO1, SNOWY COTYLEDON 1 |
-0.514 |
-0.048 |
| 68 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.498 |
-0.143 |
| 69 |
266927_at |
AT2G45960
|
ATHH2, PIP1;2, NAMED PLASMA MEMBRANE INTRINSIC PROTEIN 1;2, PIP1B, plasma membrane intrinsic protein 1B, TMP-A, TRANSMEMBRANE PROTEIN A |
-0.495 |
-0.019 |
| 70 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
-0.492 |
0.300 |
| 71 |
251962_at |
AT3G53420
|
AtPIP2;1, PIP2, PLASMA MEMBRANE INTRINSIC PROTEIN 2, PIP2;1, PLASMA MEMBRANE INTRINSIC PROTEIN 2;1, PIP2A, plasma membrane intrinsic protein 2A |
-0.489 |
-0.016 |
| 72 |
261581_at |
AT1G01140
|
CIPK9, CBL-interacting protein kinase 9, PKS6, PROTEIN KINASE 6, SnRK3.12, SNF1-RELATED PROTEIN KINASE 3.12 |
-0.484 |
0.026 |
| 73 |
260788_at |
AT1G06260
|
[Cysteine proteinases superfamily protein] |
-0.464 |
-0.056 |
| 74 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
-0.462 |
-0.023 |
| 75 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
-0.433 |
-0.043 |
| 76 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.426 |
-0.002 |
| 77 |
245483_at |
AT4G16190
|
[Papain family cysteine protease] |
-0.419 |
0.015 |
| 78 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
-0.402 |
0.060 |
| 79 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.389 |
0.356 |
| 80 |
254188_at |
AT4G23920
|
ATUGE2, UDP-GLC 4-EPIMERASE 2, UGE2, UDP-D-glucose/UDP-D-galactose 4-epimerase 2 |
-0.385 |
0.016 |
| 81 |
251324_at |
AT3G61430
|
ATPIP1, ARABIDOPSIS THALIANA PLASMA MEMBRANE INTRINSIC PROTEIN 1, PIP1, PLASMA MEMBRANE INTRINSIC PROTEIN 1, PIP1;1, PLASMA MEMBRANE INTRINSIC PROTEIN 1;1, PIP1A, plasma membrane intrinsic protein 1A |
-0.382 |
-0.052 |
| 82 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.374 |
0.025 |
| 83 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
-0.373 |
0.065 |
| 84 |
260637_at |
AT1G62380
|
ACO2, ACC oxidase 2, ATACO2 |
-0.370 |
-0.009 |
| 85 |
264143_at |
AT1G79330
|
AMC6, ATMC5, metacaspase 5, ATMCP2b, metacaspase 2b, MC5, metacaspase 5, MCP2b, metacaspase 2b |
-0.364 |
-0.011 |
| 86 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
-0.353 |
0.090 |
| 87 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.350 |
0.000 |
| 88 |
257680_at |
AT3G20330
|
PYRB, PYRIMIDINE B |
-0.347 |
0.011 |
| 89 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.345 |
-0.028 |
| 90 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.337 |
0.066 |
| 91 |
255602_at |
AT4G01026
|
PYL7, PYR1-like 7, RCAR2, regulatory components of ABA receptor 2 |
-0.333 |
0.022 |
| 92 |
254629_at |
AT4G18425
|
unknown |
-0.328 |
0.028 |
| 93 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.328 |
0.005 |
| 94 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.325 |
-0.077 |
| 95 |
265194_at |
AT1G05010
|
ACO4, ethylene forming enzyme, EAT1, EFE, ethylene-forming enzyme |
-0.324 |
-0.018 |
| 96 |
260083_at |
AT1G63220
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.324 |
0.107 |
| 97 |
252929_at |
AT4G38970
|
AtFBA2, FBA2, fructose-bisphosphate aldolase 2 |
-0.322 |
-0.025 |
| 98 |
259372_at |
AT1G69120
|
AGL7, AGAMOUS-like 7, AP1, APETALA1, AtAP1 |
-0.320 |
-0.032 |
| 99 |
265460_at |
AT2G46600
|
[Calcium-binding EF-hand family protein] |
-0.320 |
0.077 |
| 100 |
247168_at |
AT5G65860
|
[ankyrin repeat family protein] |
-0.314 |
-0.029 |
| 101 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
-0.313 |
-0.060 |
| 102 |
258140_at |
AT3G24503
|
ALDH1A, aldehyde dehydrogenase 1A, ALDH2C4, aldehyde dehydrogenase 2C4, REF1, REDUCED EPIDERMAL FLUORESCENCE1 |
-0.307 |
-0.059 |
| 103 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.307 |
-0.053 |
| 104 |
249377_at |
AT5G40690
|
unknown |
-0.307 |
-0.031 |
| 105 |
252095_at |
AT3G51000
|
[alpha/beta-Hydrolases superfamily protein] |
-0.305 |
0.010 |
| 106 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
-0.300 |
0.117 |
| 107 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.297 |
0.424 |
| 108 |
252592_at |
AT3G45640
|
ATMAPK3, ATMPK3, mitogen-activated protein kinase 3, MPK3, mitogen-activated protein kinase 3 |
-0.294 |
0.031 |
| 109 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.289 |
-0.133 |
| 110 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.289 |
0.093 |
| 111 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.288 |
-0.113 |
| 112 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.282 |
0.013 |
| 113 |
254299_at |
AT4G22920
|
ATNYE1, NON-YELLOWING 1, NYE1, NON-YELLOWING 1, SGR, STAY-GREEN |
-0.280 |
-0.070 |
| 114 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.279 |
-0.005 |
| 115 |
266458_at |
AT2G47710
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.279 |
-0.012 |