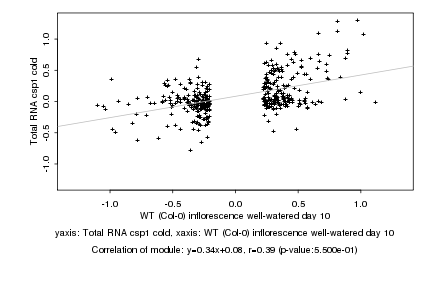
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT (Col-0) inflorescence well-watered day 10 |
Log2 signal ratio
Total RNA csp1 cold |
| 1 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
1.116 |
-0.009 |
| 2 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
1.016 |
1.084 |
| 3 |
265387_at |
AT2G20670
|
unknown |
0.995 |
0.159 |
| 4 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.967 |
1.313 |
| 5 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.891 |
0.829 |
| 6 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.889 |
0.770 |
| 7 |
244932_at |
ATCG01060
|
PSAC |
0.877 |
0.040 |
| 8 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
0.876 |
0.700 |
| 9 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
0.830 |
0.397 |
| 10 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.807 |
1.131 |
| 11 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.806 |
1.293 |
| 12 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.743 |
0.742 |
| 13 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.741 |
0.368 |
| 14 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.730 |
0.381 |
| 15 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
0.725 |
0.607 |
| 16 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.722 |
0.490 |
| 17 |
245007_at |
ATCG00350
|
PSAA |
0.681 |
-0.011 |
| 18 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.668 |
0.655 |
| 19 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.661 |
0.758 |
| 20 |
256442_at |
AT3G10930
|
unknown |
0.656 |
0.002 |
| 21 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.655 |
1.091 |
| 22 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.651 |
0.539 |
| 23 |
245025_at |
ATCG00130
|
ATPF |
0.635 |
-0.037 |
| 24 |
257085_at |
AT3G20630
|
ATUBP14, PER1, phosphate deficiency root hair defective1, TTN6, TITAN6, UBP14, ubiquitin-specific protease 14 |
0.612 |
-0.015 |
| 25 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.594 |
0.697 |
| 26 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.594 |
0.359 |
| 27 |
266990_at |
AT2G39190
|
ATATH8 |
0.568 |
-0.086 |
| 28 |
253643_at |
AT4G29780
|
unknown |
0.568 |
0.152 |
| 29 |
262211_at |
AT1G74930
|
ORA47 |
0.567 |
-0.097 |
| 30 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.565 |
0.445 |
| 31 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.556 |
-0.022 |
| 32 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.551 |
0.054 |
| 33 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.547 |
0.029 |
| 34 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.545 |
0.445 |
| 35 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
0.543 |
0.016 |
| 36 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
0.526 |
0.565 |
| 37 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.525 |
0.672 |
| 38 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.519 |
0.551 |
| 39 |
263751_at |
AT2G21300
|
[ATP binding microtubule motor family protein] |
0.517 |
-0.032 |
| 40 |
250241_at |
AT5G13600
|
[Phototropic-responsive NPH3 family protein] |
0.507 |
0.023 |
| 41 |
257709_at |
AT3G27325
|
[hydrolases, acting on ester bonds] |
0.507 |
-0.025 |
| 42 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.503 |
0.189 |
| 43 |
244965_at |
ATCG00590
|
ORF31 |
0.499 |
-0.064 |
| 44 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.492 |
0.453 |
| 45 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.481 |
-0.436 |
| 46 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.473 |
0.762 |
| 47 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.469 |
0.794 |
| 48 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
0.466 |
0.219 |
| 49 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.461 |
0.392 |
| 50 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.456 |
0.632 |
| 51 |
264518_at |
AT1G09980
|
[Putative serine esterase family protein] |
0.453 |
-0.001 |
| 52 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.447 |
0.023 |
| 53 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.444 |
0.684 |
| 54 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.444 |
0.366 |
| 55 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.444 |
0.041 |
| 56 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.444 |
0.049 |
| 57 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
0.430 |
-0.013 |
| 58 |
267511_at |
AT2G45670
|
[calcineurin B subunit-related] |
0.428 |
0.032 |
| 59 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.425 |
0.109 |
| 60 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.419 |
0.050 |
| 61 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.418 |
0.271 |
| 62 |
253605_at |
AT4G30990
|
[ARM repeat superfamily protein] |
0.415 |
0.108 |
| 63 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.413 |
0.078 |
| 64 |
261532_at |
AT1G71680
|
[Transmembrane amino acid transporter family protein] |
0.411 |
0.007 |
| 65 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.411 |
0.580 |
| 66 |
250595_at |
AT5G07770
|
AtFH16, FH16, formin homolog 16 |
0.411 |
-0.078 |
| 67 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.409 |
0.759 |
| 68 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
0.406 |
0.248 |
| 69 |
255640_at |
AT4G00800
|
SETH5 |
0.402 |
-0.046 |
| 70 |
249209_at |
AT5G42620
|
[metalloendopeptidases] |
0.393 |
-0.073 |
| 71 |
257850_at |
AT3G13065
|
SRF4, STRUBBELIG-receptor family 4 |
0.393 |
0.009 |
| 72 |
244966_at |
ATCG00600
|
PETG |
0.392 |
-0.041 |
| 73 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
0.390 |
0.216 |
| 74 |
263799_at |
AT2G24550
|
unknown |
0.388 |
0.053 |
| 75 |
246896_at |
AT5G25550
|
[Leucine-rich repeat (LRR) family protein] |
0.387 |
0.011 |
| 76 |
245587_at |
AT4G15020
|
[hAT transposon superfamily] |
0.383 |
0.053 |
| 77 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.382 |
-0.043 |
| 78 |
247585_at |
AT5G60680
|
unknown |
0.380 |
0.272 |
| 79 |
245897_at |
AT5G09400
|
KUP7, K+ uptake permease 7 |
0.380 |
0.039 |
| 80 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.379 |
0.081 |
| 81 |
263384_at |
AT2G40130
|
SMXL8, SMAX1-like 8 |
0.377 |
0.139 |
| 82 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.377 |
0.067 |
| 83 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.370 |
0.088 |
| 84 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.370 |
0.494 |
| 85 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.367 |
0.387 |
| 86 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.367 |
0.518 |
| 87 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
0.364 |
0.132 |
| 88 |
266583_at |
AT2G46220
|
[Uncharacterized conserved protein (DUF2358)] |
0.363 |
-0.040 |
| 89 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.360 |
0.560 |
| 90 |
255036_at |
AT4G09560
|
[Protease-associated (PA) RING/U-box zinc finger family protein] |
0.357 |
0.186 |
| 91 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.355 |
0.936 |
| 92 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.355 |
-0.103 |
| 93 |
244962_at |
ATCG01050
|
NDHD |
0.351 |
-0.059 |
| 94 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.351 |
0.491 |
| 95 |
245018_at |
ATCG00520
|
YCF4 |
0.347 |
-0.043 |
| 96 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
0.343 |
-0.021 |
| 97 |
264684_at |
AT1G65590
|
ATHEX1, HEXO3, beta-hexosaminidase 3 |
0.342 |
0.018 |
| 98 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.342 |
0.528 |
| 99 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.339 |
0.167 |
| 100 |
258985_at |
AT3G08960
|
[ARM repeat superfamily protein] |
0.339 |
-0.011 |
| 101 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.338 |
0.391 |
| 102 |
249761_at |
AT5G23970
|
[HXXXD-type acyl-transferase family protein] |
0.338 |
0.024 |
| 103 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
0.332 |
0.076 |
| 104 |
245290_at |
AT4G16490
|
[ARM repeat superfamily protein] |
0.330 |
0.027 |
| 105 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.330 |
0.193 |
| 106 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
0.329 |
0.243 |
| 107 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.329 |
0.529 |
| 108 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.327 |
0.600 |
| 109 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.326 |
-0.097 |
| 110 |
262463_at |
AT1G50310
|
ATSTP9, SUGAR TRANSPORTER 9, STP9, sugar transporter 9 |
0.326 |
0.131 |
| 111 |
255673_at |
AT4G00340
|
RLK4, receptor-like protein kinase 4 |
0.326 |
0.064 |
| 112 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.325 |
0.087 |
| 113 |
247408_at |
AT5G62760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.324 |
-0.203 |
| 114 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.323 |
0.040 |
| 115 |
259015_at |
AT3G07350
|
unknown |
0.323 |
0.087 |
| 116 |
260010_at |
AT1G68020
|
ATTPS6, TPS6, TREHALOSE -6-PHOSPHATASE SYNTHASE S6 |
0.322 |
0.047 |
| 117 |
253859_at |
AT4G27657
|
unknown |
0.321 |
0.338 |
| 118 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.321 |
0.863 |
| 119 |
256896_at |
AT3G24630
|
TRM34, TON1 Recruiting Motif 34 |
0.315 |
0.093 |
| 120 |
253881_at |
AT4G27640
|
[ARM repeat superfamily protein] |
0.314 |
-0.010 |
| 121 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.314 |
0.503 |
| 122 |
245490_at |
AT4G16310
|
LDL3, LSD1-like 3 |
0.313 |
0.035 |
| 123 |
265767_at |
AT2G48110
|
MED33B, REF4, REDUCED EPIDERMAL FLUORESCENCE 4 |
0.311 |
-0.002 |
| 124 |
252207_at |
AT3G50370
|
unknown |
0.311 |
-0.038 |
| 125 |
256262_at |
AT3G12150
|
unknown |
0.309 |
0.042 |
| 126 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.309 |
0.482 |
| 127 |
254178_at |
AT4G23880
|
unknown |
0.309 |
0.309 |
| 128 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
0.309 |
-0.042 |
| 129 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.307 |
0.010 |
| 130 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
0.306 |
0.090 |
| 131 |
266019_at |
AT2G18750
|
[Calmodulin-binding protein] |
0.305 |
0.097 |
| 132 |
247025_at |
AT5G67030
|
ABA1, ABA DEFICIENT 1, ATABA1, ARABIDOPSIS THALIANA ABA DEFICIENT 1, ATZEP, ARABIDOPSIS THALIANA ZEAXANTHIN EPOXIDASE, IBS3, IMPAIRED IN BABA-INDUCED STERILITY 3, LOS6, LOW EXPRESSION OF OSMOTIC STRESS-RESPONSIVE GENES 6, NPQ2, NON-PHOTOCHEMICAL QUENCHING 2, ZEP, ZEAXANTHIN EPOXIDASE |
0.303 |
0.285 |
| 133 |
244967_at |
ATCG00630
|
PSAJ |
0.303 |
-0.111 |
| 134 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
0.301 |
0.536 |
| 135 |
260045_at |
AT1G73670
|
ATMPK15, MAP kinase 15, MPK15, MAP kinase 15 |
0.301 |
0.016 |
| 136 |
253305_at |
AT4G33666
|
unknown |
0.300 |
-0.476 |
| 137 |
245019_at |
ATCG00530
|
YCF10 |
0.300 |
-0.025 |
| 138 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.297 |
-0.120 |
| 139 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.297 |
0.448 |
| 140 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
0.290 |
0.581 |
| 141 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.286 |
0.658 |
| 142 |
258382_at |
AT3G15355
|
PFU1, PHO2 FAMILY UBIQUITIN CONJUGATION ENZYME 1, UBC25, ubiquitin-conjugating enzyme 25 |
0.286 |
-0.071 |
| 143 |
260032_at |
AT1G68750
|
ATPPC4, phosphoenolpyruvate carboxylase 4, PPC4, phosphoenolpyruvate carboxylase 4 |
0.286 |
-0.028 |
| 144 |
262585_at |
AT1G15460
|
ATBOR4, ARABIDOPSIS THALIANA REQUIRES HIGH BORON 4, BOR4, REQUIRES HIGH BORON 4 |
0.286 |
-0.034 |
| 145 |
254581_at |
AT4G19440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.285 |
0.045 |
| 146 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.282 |
0.455 |
| 147 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.281 |
0.507 |
| 148 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.277 |
0.304 |
| 149 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.276 |
0.027 |
| 150 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
0.275 |
0.202 |
| 151 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.272 |
0.221 |
| 152 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.270 |
-0.070 |
| 153 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.268 |
-0.018 |
| 154 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.267 |
0.041 |
| 155 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.265 |
-0.086 |
| 156 |
263931_at |
AT2G36220
|
unknown |
0.265 |
0.470 |
| 157 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.263 |
0.311 |
| 158 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
0.262 |
-0.080 |
| 159 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.261 |
0.142 |
| 160 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.261 |
0.229 |
| 161 |
256721_at |
AT2G34150
|
ATRANGAP2, ATSCAR1, SCAR1, WAVE1, WISKOTT-ALDRICH SYNDROME PROTEIN FAMILY VERPROLIN HOMOLOGOUS PROTEIN 1 |
0.260 |
-0.305 |
| 162 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
0.258 |
0.066 |
| 163 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.258 |
0.585 |
| 164 |
247232_at |
AT5G64940
|
ATATH13, ARABIDOPSIS THALIANA ABC2 HOMOLOG 13, ATH13, ABC2 homolog 13, ATOSA1, A. THALIANA OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1, OSA1, OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1 |
0.256 |
0.142 |
| 165 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.256 |
0.237 |
| 166 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.256 |
0.019 |
| 167 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.255 |
0.350 |
| 168 |
259017_at |
AT3G07310
|
unknown |
0.254 |
-0.077 |
| 169 |
254111_at |
AT4G24890
|
ATPAP24, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 24, PAP24, purple acid phosphatase 24 |
0.253 |
-0.026 |
| 170 |
265731_at |
AT2G01130
|
[DEA(D/H)-box RNA helicase family protein] |
0.252 |
-0.026 |
| 171 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.251 |
0.361 |
| 172 |
249134_at |
AT5G43150
|
unknown |
0.250 |
0.163 |
| 173 |
258461_at |
AT3G17340
|
[ARM repeat superfamily protein] |
0.248 |
-0.065 |
| 174 |
247693_at |
AT5G59730
|
ATEXO70H7, exocyst subunit exo70 family protein H7, EXO70H7, exocyst subunit exo70 family protein H7 |
0.246 |
0.292 |
| 175 |
245016_at |
ATCG00500
|
ACCD, acetyl-CoA carboxylase carboxyl transferase subunit beta |
0.246 |
-0.051 |
| 176 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.245 |
0.434 |
| 177 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
0.245 |
0.395 |
| 178 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.244 |
0.629 |
| 179 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.243 |
0.253 |
| 180 |
247928_at |
AT5G57320
|
VLN5, villin 5 |
0.243 |
0.002 |
| 181 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.243 |
0.295 |
| 182 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.241 |
-0.103 |
| 183 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.240 |
0.935 |
| 184 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
0.239 |
0.378 |
| 185 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.239 |
0.021 |
| 186 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.238 |
0.124 |
| 187 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
0.238 |
-0.003 |
| 188 |
257407_at |
AT1G27100
|
[Actin cross-linking protein] |
0.237 |
0.032 |
| 189 |
249905_at |
AT5G22720
|
[F-box/RNI-like superfamily protein] |
0.235 |
0.219 |
| 190 |
262949_at |
AT1G75790
|
sks18, SKU5 similar 18 |
0.232 |
-0.044 |
| 191 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.232 |
0.294 |
| 192 |
264238_at |
AT1G54740
|
unknown |
0.231 |
0.006 |
| 193 |
263163_at |
AT1G03160
|
FZL, FZO-like |
0.231 |
-0.210 |
| 194 |
267346_at |
AT2G39940
|
COI1, CORONATINE INSENSITIVE 1 |
0.231 |
0.013 |
| 195 |
257205_at |
AT3G16520
|
UGT88A1, UDP-glucosyl transferase 88A1 |
0.229 |
-0.061 |
| 196 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
0.228 |
0.289 |
| 197 |
257321_at |
ATMG01130
|
ORF106F |
0.227 |
-0.038 |
| 198 |
251025_at |
AT5G02190
|
ATASP38, ARABIDOPSIS THALIANA ASPARTIC PROTEASE 38, EMB24, EMBRYO DEFECTIVE 24, PCS1, PROMOTION OF CELL SURVIVAL 1 |
0.227 |
0.037 |
| 199 |
251814_at |
AT3G54890
|
LHCA1, photosystem I light harvesting complex gene 1 |
0.227 |
0.068 |
| 200 |
250078_at |
AT5G16690
|
ATORC3, ORC3, origin recognition complex subunit 3 |
0.226 |
-0.058 |
| 201 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
0.225 |
0.243 |
| 202 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.224 |
0.611 |
| 203 |
257349_at |
AT2G30630
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.223 |
0.060 |
| 204 |
263962_at |
AT2G36350
|
[Protein kinase superfamily protein] |
0.223 |
0.231 |
| 205 |
257589_at |
AT1G55050
|
unknown |
0.223 |
0.064 |
| 206 |
250232_at |
AT5G13950
|
unknown |
0.222 |
0.193 |
| 207 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.221 |
0.048 |
| 208 |
267545_at |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
-1.102 |
-0.062 |
| 209 |
258289_at |
AT3G23450
|
unknown |
-1.054 |
-0.071 |
| 210 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-1.038 |
-0.119 |
| 211 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.994 |
0.358 |
| 212 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.986 |
-0.444 |
| 213 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.964 |
-0.484 |
| 214 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.938 |
0.009 |
| 215 |
253532_at |
AT4G31570
|
unknown |
-0.856 |
-0.038 |
| 216 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.828 |
-0.346 |
| 217 |
248350_at |
AT5G52160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.790 |
-0.199 |
| 218 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.787 |
0.062 |
| 219 |
261684_at |
AT1G47400
|
unknown |
-0.784 |
-0.621 |
| 220 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
-0.713 |
-0.218 |
| 221 |
248862_at |
AT5G46730
|
[glycine-rich protein] |
-0.702 |
0.072 |
| 222 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
-0.683 |
-0.017 |
| 223 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.650 |
-0.030 |
| 224 |
250828_at |
AT5G05250
|
unknown |
-0.618 |
-0.578 |
| 225 |
264430_at |
AT1G61680
|
ATTPS14, TERPENE SYNTHASE 14, TPS14, terpene synthase 14 |
-0.584 |
0.009 |
| 226 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.584 |
-0.012 |
| 227 |
249708_at |
AT5G35660
|
[Glycine-rich protein family] |
-0.574 |
0.081 |
| 228 |
253322_at |
AT4G33980
|
unknown |
-0.573 |
0.303 |
| 229 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.561 |
0.028 |
| 230 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.561 |
0.269 |
| 231 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.547 |
-0.392 |
| 232 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.545 |
0.256 |
| 233 |
260688_at |
AT1G17665
|
unknown |
-0.544 |
0.349 |
| 234 |
267036_at |
AT2G38465
|
unknown |
-0.539 |
0.270 |
| 235 |
265567_at |
AT2G05580
|
[Glycine-rich protein family] |
-0.533 |
0.067 |
| 236 |
261764_at |
AT1G15530
|
LecRK-S.1, L-type lectin receptor kinase S.1 |
-0.522 |
0.025 |
| 237 |
245700_at |
AT5G04180
|
ACA3, alpha carbonic anhydrase 3, ATACA3, ALPHA CARBONIC ANHYDRASE 3 |
-0.520 |
-0.023 |
| 238 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.514 |
-0.080 |
| 239 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.511 |
-0.195 |
| 240 |
261946_at |
AT1G64560
|
|
-0.506 |
-0.049 |
| 241 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.505 |
0.145 |
| 242 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
-0.485 |
-0.378 |
| 243 |
266358_at |
AT2G32280
|
VCC, VASCULATURE COMPLEXITY AND CONNECTIVITY |
-0.482 |
0.367 |
| 244 |
245008_at |
ATCG00360
|
YCF3 |
-0.480 |
-0.027 |
| 245 |
267560_at |
AT2G45580
|
CYP76C3, cytochrome P450, family 76, subfamily C, polypeptide 3 |
-0.466 |
0.053 |
| 246 |
249357_at |
AT5G40490
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.465 |
0.111 |
| 247 |
245486_x_at |
AT4G16240
|
unknown |
-0.463 |
0.015 |
| 248 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
-0.460 |
0.071 |
| 249 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.444 |
0.283 |
| 250 |
263144_at |
AT1G54070
|
[Dormancy/auxin associated family protein] |
-0.443 |
0.042 |
| 251 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.440 |
-0.004 |
| 252 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.440 |
-0.434 |
| 253 |
253082_at |
AT4G36230
|
unknown |
-0.412 |
0.076 |
| 254 |
259692_at |
AT1G63080
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.411 |
0.060 |
| 255 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.409 |
0.209 |
| 256 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.408 |
-0.110 |
| 257 |
260905_at |
AT1G02710
|
[glycine-rich protein] |
-0.407 |
0.040 |
| 258 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.395 |
0.202 |
| 259 |
259363_at |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
-0.386 |
-0.014 |
| 260 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.386 |
-0.007 |
| 261 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
-0.385 |
-0.152 |
| 262 |
257331_at |
ATMG01330
|
ORF107H |
-0.385 |
0.039 |
| 263 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.376 |
0.015 |
| 264 |
265448_at |
AT2G46590
|
Dof-type zinc finger DNA-binding family protein |
-0.376 |
0.102 |
| 265 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.375 |
-0.030 |
| 266 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.371 |
-0.058 |
| 267 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.371 |
0.365 |
| 268 |
258358_at |
AT3G14380
|
[Uncharacterised protein family (UPF0497)] |
-0.371 |
-0.051 |
| 269 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
-0.366 |
0.316 |
| 270 |
248257_at |
AT5G53410
|
unknown |
-0.363 |
-0.030 |
| 271 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.363 |
-0.775 |
| 272 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
-0.360 |
0.299 |
| 273 |
246039_at |
AT5G19480
|
unknown |
-0.357 |
-0.091 |
| 274 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.352 |
-0.054 |
| 275 |
256824_at |
AT3G22121
|
other RNA |
-0.347 |
0.031 |
| 276 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.345 |
-0.331 |
| 277 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.342 |
-0.120 |
| 278 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.340 |
-0.433 |
| 279 |
244903_at |
ATMG00660
|
ORF149 |
-0.338 |
0.018 |
| 280 |
262892_at |
AT1G79440
|
ALDH5F1, aldehyde dehydrogenase 5F1, ENF1, ENLARGED FIL EXPRESSION DOMAIN 1, SSADH, SUCCINIC SEMIALDEHYDE DEHYDROGENASE, SSADH1, SUCCINIC SEMIALDEHYDE DEHYDROGENASE 1 |
-0.334 |
-0.068 |
| 281 |
259031_at |
AT3G09360
|
[Cyclin/Brf1-like TBP-binding protein] |
-0.333 |
0.029 |
| 282 |
265803_at |
AT2G18115
|
[pseudogene] |
-0.333 |
-0.034 |
| 283 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.332 |
0.172 |
| 284 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
-0.331 |
-0.037 |
| 285 |
256603_at |
AT3G28270
|
unknown |
-0.330 |
-0.314 |
| 286 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
-0.327 |
0.109 |
| 287 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
-0.322 |
0.103 |
| 288 |
259493_at |
AT1G15840
|
unknown |
-0.318 |
0.077 |
| 289 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.318 |
-0.090 |
| 290 |
260689_at |
AT1G32290
|
unknown |
-0.313 |
0.078 |
| 291 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
-0.312 |
-0.346 |
| 292 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.312 |
0.560 |
| 293 |
261528_at |
AT1G14420
|
AT59 |
-0.311 |
0.009 |
| 294 |
258108_at |
AT3G23570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.311 |
-0.104 |
| 295 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
-0.310 |
0.104 |
| 296 |
254629_at |
AT4G18425
|
unknown |
-0.308 |
-0.152 |
| 297 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.308 |
-0.155 |
| 298 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-0.307 |
-0.041 |
| 299 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.307 |
-0.231 |
| 300 |
260674_at |
AT1G19370
|
unknown |
-0.306 |
0.278 |
| 301 |
246525_at |
AT5G15840
|
BBX1, B-box domain protein 1, CO, CONSTANS, FG |
-0.306 |
0.215 |
| 302 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.305 |
-0.015 |
| 303 |
263775_at |
AT2G46410
|
CPC, CAPRICE |
-0.304 |
-0.050 |
| 304 |
257874_at |
AT3G17110
|
[pseudogene] |
-0.304 |
-0.021 |
| 305 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.304 |
-0.330 |
| 306 |
258748_at |
AT3G05930
|
GLP8, germin-like protein 8 |
-0.302 |
-0.060 |
| 307 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.302 |
0.064 |
| 308 |
248307_at |
AT5G52850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.301 |
-0.149 |
| 309 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
-0.300 |
0.677 |
| 310 |
249505_at |
AT5G38870
|
[pseudogene] |
-0.299 |
0.030 |
| 311 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.298 |
0.394 |
| 312 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.297 |
-0.030 |
| 313 |
262236_at |
AT1G48330
|
unknown |
-0.296 |
-0.462 |
| 314 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
-0.293 |
-0.364 |
| 315 |
261147_at |
AT1G19690
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.292 |
-0.215 |
| 316 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.291 |
-0.255 |
| 317 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
-0.288 |
0.035 |
| 318 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
-0.287 |
0.155 |
| 319 |
252627_at |
AT3G44950
|
[glycine-rich protein] |
-0.286 |
0.127 |
| 320 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.285 |
-0.373 |
| 321 |
249404_at |
AT5G40160
|
EMB139, EMBRYO DEFECTIVE 139, EMB506, embryo defective 506 |
-0.283 |
-0.052 |
| 322 |
251292_at |
AT3G61920
|
unknown |
-0.283 |
-0.380 |
| 323 |
251704_at |
AT3G56360
|
unknown |
-0.283 |
-0.248 |
| 324 |
257619_at |
AT3G24810
|
ICK3, KRP5, kip-related protein 5 |
-0.283 |
-0.101 |
| 325 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.281 |
-0.018 |
| 326 |
252786_at |
AT3G42670
|
CHR38, chromatin remodeling 38, CLSY, CLASSY1, CLSY1, CLASSY 1 |
-0.280 |
-0.125 |
| 327 |
259928_at |
AT1G34380
|
[5'-3' exonuclease family protein] |
-0.276 |
-0.117 |
| 328 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.275 |
-0.013 |
| 329 |
251223_at |
AT3G62610
|
ATMYB11, myb domain protein 11, MYB11, myb domain protein 11, PFG2, PRODUCTION OF FLAVONOL GLYCOSIDES 2 |
-0.271 |
-0.030 |
| 330 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.271 |
-0.106 |
| 331 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.271 |
-0.650 |
| 332 |
264026_at |
AT2G21060
|
ATCSP4, COLD SHOCK DOMAIN PROTEIN 4, ATGRP2B, glycine-rich protein 2B, GRP2B, glycine-rich protein 2B |
-0.270 |
0.093 |
| 333 |
258951_at |
AT3G01380
|
[transferases] |
-0.270 |
0.023 |
| 334 |
260332_at |
AT1G70470
|
unknown |
-0.269 |
-0.106 |
| 335 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
-0.269 |
-0.075 |
| 336 |
247550_at |
AT5G61370
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.267 |
0.266 |
| 337 |
247937_at |
AT5G57110
|
ACA8, autoinhibited Ca2+ -ATPase, isoform 8, AT-ACA8, AUTOINHIBITED CA2+ -ATPASE, ISOFORM 8 |
-0.265 |
0.317 |
| 338 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.263 |
-0.045 |
| 339 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
-0.259 |
0.075 |
| 340 |
253317_at |
AT4G33960
|
unknown |
-0.257 |
-0.272 |
| 341 |
249995_at |
AT5G18610
|
[Protein kinase superfamily protein] |
-0.257 |
0.018 |
| 342 |
257685_at |
AT3G12770
|
MEF22, mitochondrial editing factor 22 |
-0.256 |
-0.077 |
| 343 |
252198_x_at |
AT3G50250
|
unknown |
-0.255 |
0.009 |
| 344 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.255 |
-0.056 |
| 345 |
258245_at |
AT3G29075
|
[glycine-rich protein] |
-0.252 |
-0.058 |
| 346 |
249515_at |
AT5G38530
|
TSBtype2, tryptophan synthase beta type 2 |
-0.252 |
-0.284 |
| 347 |
267476_at |
AT2G02720
|
[Pectate lyase family protein] |
-0.251 |
0.125 |
| 348 |
254610_at |
AT4G18890
|
BEH3, BES1/BZR1 homolog 3 |
-0.251 |
-0.092 |
| 349 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.248 |
-0.029 |
| 350 |
260883_at |
AT1G29270
|
unknown |
-0.245 |
0.314 |
| 351 |
267636_at |
AT2G42110
|
unknown |
-0.245 |
0.103 |
| 352 |
246587_at |
AT5G14830
|
[retrotransposon family] |
-0.244 |
0.155 |
| 353 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.244 |
0.065 |
| 354 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.243 |
-0.083 |
| 355 |
256856_at |
AT3G15110
|
unknown |
-0.242 |
-0.086 |
| 356 |
263409_at |
AT2G04063
|
[glycine-rich protein] |
-0.241 |
-0.115 |
| 357 |
266673_at |
AT2G29630
|
PY, PYRIMIDINE REQUIRING, THIC, thiaminC |
-0.239 |
-0.185 |
| 358 |
252117_at |
AT3G51430
|
SSL5, STRICTOSIDINE SYNTHASE-LIKE 5, YLS2, YELLOW-LEAF-SPECIFIC GENE 2 |
-0.239 |
-0.376 |
| 359 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
-0.238 |
0.057 |
| 360 |
266325_at |
AT2G46630
|
unknown |
-0.237 |
-0.132 |
| 361 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.236 |
-0.246 |
| 362 |
252110_at |
AT3G51560
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.236 |
0.043 |
| 363 |
252210_at |
AT3G50410
|
OBP1, OBF binding protein 1 |
-0.235 |
0.224 |
| 364 |
250360_at |
AT5G11360
|
[Interleukin-1 receptor-associated kinase 4 protein] |
-0.235 |
0.030 |
| 365 |
247567_at |
AT5G61190
|
[putative endonuclease or glycosyl hydrolase with C2H2-type zinc finger domain] |
-0.234 |
-0.275 |
| 366 |
254756_at |
AT4G13235
|
EDA21, embryo sac development arrest 21 |
-0.234 |
-0.071 |
| 367 |
253292_at |
AT4G33985
|
unknown |
-0.234 |
-0.026 |
| 368 |
248538_at |
AT5G50110
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.232 |
-0.061 |
| 369 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
-0.232 |
-0.047 |
| 370 |
261805_at |
AT1G30540
|
[Actin-like ATPase superfamily protein] |
-0.231 |
-0.070 |
| 371 |
267193_at |
AT2G30900
|
TBL43, TRICHOME BIREFRINGENCE-LIKE 43 |
-0.230 |
-0.039 |
| 372 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.230 |
0.092 |
| 373 |
264673_at |
AT1G09795
|
ATATP-PRT2, ATP phosphoribosyl transferase 2, ATP-PRT2, ATP phosphoribosyl transferase 2, HISN1B |
-0.230 |
-0.080 |
| 374 |
251166_at |
AT3G63350
|
AT-HSFA7B, HSFA7B, HEAT SHOCK TRANSCRIPTION FACTOR A7B |
-0.229 |
0.012 |
| 375 |
250391_at |
AT5G10850
|
[similar to nucleic acid binding / zinc ion binding [Arabidopsis thaliana] (TAIR:AT2G01050.1)] |
-0.229 |
0.161 |
| 376 |
261155_at |
AT1G04960
|
unknown |
-0.229 |
0.054 |
| 377 |
247319_at |
AT5G64050
|
ATERS, ERS, glutamate tRNA synthetase, OVA3, OVULE ABORTION 3 |
-0.228 |
0.035 |
| 378 |
256674_at |
AT3G52360
|
unknown |
-0.228 |
-0.569 |
| 379 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
-0.228 |
-0.034 |
| 380 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.227 |
-0.240 |
| 381 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
-0.227 |
0.053 |
| 382 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
-0.227 |
0.245 |
| 383 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.227 |
0.204 |
| 384 |
262338_at |
AT1G64185
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.227 |
-0.130 |
| 385 |
259299_at |
AT3G05080
|
unknown |
-0.227 |
-0.061 |
| 386 |
264879_at |
AT1G61260
|
unknown |
-0.226 |
-0.143 |
| 387 |
248097_at |
AT5G55260
|
PPX-2, PROTEIN PHOSPHATASE X -2, PPX2, protein phosphatase X 2 |
-0.226 |
-0.075 |
| 388 |
254842_at |
AT4G11950
|
unknown |
-0.226 |
0.017 |
| 389 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.226 |
-0.360 |
| 390 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.224 |
-0.071 |
| 391 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
-0.223 |
-0.257 |
| 392 |
248228_at |
AT5G53800
|
unknown |
-0.222 |
-0.052 |
| 393 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.222 |
-0.019 |
| 394 |
249574_at |
AT5G37660
|
PDLP7, plasmodesmata-located protein 7 |
-0.221 |
-0.078 |
| 395 |
247594_at |
AT5G60800
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.220 |
-0.314 |
| 396 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
-0.220 |
0.003 |
| 397 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.219 |
-0.022 |
| 398 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.219 |
-0.326 |
| 399 |
251110_at |
AT5G01260
|
[Carbohydrate-binding-like fold] |
-0.218 |
-0.005 |
| 400 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.216 |
-0.037 |
| 401 |
247950_at |
AT5G57230
|
[Thioredoxin superfamily protein] |
-0.215 |
-0.060 |
| 402 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.214 |
-0.136 |
| 403 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
-0.214 |
-0.019 |
| 404 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.214 |
-0.041 |
| 405 |
256255_at |
AT3G11280
|
[Duplicated homeodomain-like superfamily protein] |
-0.214 |
0.018 |
| 406 |
250956_at |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
-0.214 |
-0.042 |
| 407 |
263681_at |
AT1G26840
|
ATORC6, ARABIDOPSIS THALIANA ORIGIN RECOGNITION COMPLEX PROTEIN 6, ORC6, origin recognition complex protein 6 |
-0.213 |
0.208 |
| 408 |
253297_at |
AT4G33810
|
[Glycosyl hydrolase superfamily protein] |
-0.213 |
0.090 |
| 409 |
256683_at |
AT3G52220
|
unknown |
-0.212 |
0.008 |
| 410 |
245401_at |
AT4G17670
|
unknown |
-0.212 |
0.274 |
| 411 |
262693_at |
AT1G62780
|
unknown |
-0.212 |
-0.028 |
| 412 |
257483_at |
AT1G49620
|
AtKRP7, ICK5, ICN6, KRP7, KIP-RELATED PROTEIN 7 |
-0.212 |
-0.123 |
| 413 |
254245_at |
AT4G23240
|
CRK16, cysteine-rich RLK (RECEPTOR-like protein kinase) 16 |
-0.212 |
-0.224 |