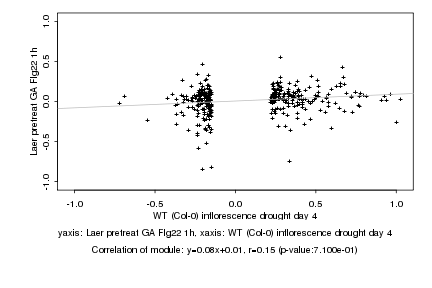
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT (Col-0) inflorescence drought day 4 |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
1.024 |
0.027 |
| 2 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
1.001 |
-0.255 |
| 3 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.961 |
0.089 |
| 4 |
245007_at |
ATCG00350
|
PSAA |
0.936 |
0.020 |
| 5 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.926 |
0.066 |
| 6 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.904 |
0.020 |
| 7 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.811 |
0.072 |
| 8 |
256442_at |
AT3G10930
|
unknown |
0.794 |
0.087 |
| 9 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.775 |
0.103 |
| 10 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.769 |
-0.059 |
| 11 |
253643_at |
AT4G29780
|
unknown |
0.769 |
0.070 |
| 12 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.765 |
-0.040 |
| 13 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.744 |
0.118 |
| 14 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.742 |
0.118 |
| 15 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.725 |
-0.126 |
| 16 |
246270_at |
AT4G36500
|
unknown |
0.720 |
0.051 |
| 17 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.719 |
0.071 |
| 18 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.687 |
0.102 |
| 19 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.677 |
-0.122 |
| 20 |
256021_at |
AT1G58270
|
ZW9 |
0.672 |
0.219 |
| 21 |
264774_at |
AT1G22890
|
unknown |
0.669 |
0.312 |
| 22 |
264518_at |
AT1G09980
|
[Putative serine esterase family protein] |
0.664 |
0.432 |
| 23 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.653 |
0.235 |
| 24 |
257085_at |
AT3G20630
|
ATUBP14, PER1, phosphate deficiency root hair defective1, TTN6, TITAN6, UBP14, ubiquitin-specific protease 14 |
0.649 |
0.189 |
| 25 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.646 |
-0.082 |
| 26 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.625 |
0.198 |
| 27 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.620 |
-0.020 |
| 28 |
266990_at |
AT2G39190
|
ATATH8 |
0.596 |
-0.325 |
| 29 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.595 |
0.156 |
| 30 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.578 |
-0.056 |
| 31 |
247061_at |
AT5G66780
|
unknown |
0.575 |
0.098 |
| 32 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.573 |
-0.001 |
| 33 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.564 |
0.052 |
| 34 |
265387_at |
AT2G20670
|
unknown |
0.563 |
0.046 |
| 35 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
0.555 |
-0.131 |
| 36 |
253881_at |
AT4G27640
|
[ARM repeat superfamily protein] |
0.538 |
0.002 |
| 37 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.527 |
-0.107 |
| 38 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.516 |
0.069 |
| 39 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.515 |
0.124 |
| 40 |
250676_at |
AT5G06320
|
NHL3, NDR1/HIN1-like 3 |
0.512 |
0.200 |
| 41 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.509 |
0.275 |
| 42 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.508 |
0.067 |
| 43 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.498 |
0.060 |
| 44 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.496 |
0.086 |
| 45 |
263605_at |
AT2G16485
|
NERD, Needed for RDR2-independent DNA methylation |
0.493 |
0.057 |
| 46 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.488 |
0.037 |
| 47 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.474 |
0.009 |
| 48 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
0.467 |
0.315 |
| 49 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.466 |
-0.224 |
| 50 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.465 |
-0.021 |
| 51 |
263931_at |
AT2G36220
|
unknown |
0.459 |
0.187 |
| 52 |
255640_at |
AT4G00800
|
SETH5 |
0.449 |
0.004 |
| 53 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
0.432 |
0.031 |
| 54 |
257709_at |
AT3G27325
|
[hydrolases, acting on ester bonds] |
0.430 |
0.147 |
| 55 |
257076_at |
AT3G19680
|
unknown |
0.430 |
-0.088 |
| 56 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.426 |
-0.284 |
| 57 |
244967_at |
ATCG00630
|
PSAJ |
0.422 |
0.033 |
| 58 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.416 |
-0.011 |
| 59 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
0.416 |
-0.003 |
| 60 |
255673_at |
AT4G00340
|
RLK4, receptor-like protein kinase 4 |
0.414 |
-0.033 |
| 61 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.411 |
-0.065 |
| 62 |
267511_at |
AT2G45670
|
[calcineurin B subunit-related] |
0.409 |
0.084 |
| 63 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.405 |
0.033 |
| 64 |
264480_at |
AT1G77300
|
ASHH2, ASH1 HOMOLOG 2, CCR1, CAROTENOID CHLOROPLAST REGULATORY1, EFS, EARLY FLOWERING IN SHORT DAYS, LAZ2, LAZARUS 2, SDG8, SET DOMAIN GROUP 8 |
0.396 |
0.130 |
| 65 |
252592_at |
AT3G45640
|
ATMAPK3, ATMPK3, mitogen-activated protein kinase 3, MPK3, mitogen-activated protein kinase 3 |
0.395 |
-0.029 |
| 66 |
261193_at |
AT1G32920
|
unknown |
0.394 |
0.030 |
| 67 |
245765_at |
AT1G33600
|
[Leucine-rich repeat (LRR) family protein] |
0.391 |
0.072 |
| 68 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.390 |
0.079 |
| 69 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.386 |
0.044 |
| 70 |
253284_at |
AT4G34150
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.386 |
0.153 |
| 71 |
262199_at |
AT1G53800
|
unknown |
0.384 |
-0.211 |
| 72 |
249209_at |
AT5G42620
|
[metalloendopeptidases] |
0.383 |
0.132 |
| 73 |
261446_at |
AT1G21170
|
SEC5B |
0.383 |
0.253 |
| 74 |
245095_at |
AT2G40860
|
[protein kinase family protein / protein phosphatase 2C ( PP2C) family protein] |
0.382 |
-0.144 |
| 75 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.381 |
-0.040 |
| 76 |
263384_at |
AT2G40130
|
SMXL8, SMAX1-like 8 |
0.380 |
0.136 |
| 77 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.380 |
-0.000 |
| 78 |
257670_at |
AT3G20340
|
unknown |
0.372 |
0.209 |
| 79 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.370 |
0.111 |
| 80 |
246896_at |
AT5G25550
|
[Leucine-rich repeat (LRR) family protein] |
0.367 |
0.062 |
| 81 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.365 |
-0.054 |
| 82 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.359 |
0.052 |
| 83 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
0.359 |
0.041 |
| 84 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
0.358 |
0.082 |
| 85 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.357 |
0.116 |
| 86 |
253605_at |
AT4G30990
|
[ARM repeat superfamily protein] |
0.355 |
0.067 |
| 87 |
247585_at |
AT5G60680
|
unknown |
0.354 |
0.019 |
| 88 |
261405_at |
AT1G18740
|
unknown |
0.353 |
0.066 |
| 89 |
265767_at |
AT2G48110
|
MED33B, REF4, REDUCED EPIDERMAL FLUORESCENCE 4 |
0.351 |
-0.013 |
| 90 |
253152_at |
AT4G35690
|
unknown |
0.348 |
0.067 |
| 91 |
251981_at |
AT3G53080
|
[D-galactoside/L-rhamnose binding SUEL lectin protein] |
0.348 |
0.074 |
| 92 |
252140_at |
AT3G51070
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.341 |
0.077 |
| 93 |
248969_at |
AT5G45310
|
unknown |
0.340 |
-0.357 |
| 94 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.334 |
-0.745 |
| 95 |
258465_at |
AT3G17220
|
ATPMEI2, PMEI2, pectin methylesterase inhibitor 2 |
0.332 |
0.092 |
| 96 |
257726_at |
AT3G18360
|
[VQ motif-containing protein] |
0.331 |
0.096 |
| 97 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.331 |
0.234 |
| 98 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.328 |
-0.018 |
| 99 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.328 |
0.127 |
| 100 |
260282_at |
AT1G80410
|
EMB2753, EMBRYO DEFECTIVE 2753, OMA, OMISHA |
0.328 |
0.050 |
| 101 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.327 |
0.151 |
| 102 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.326 |
0.072 |
| 103 |
244966_at |
ATCG00600
|
PETG |
0.325 |
-0.052 |
| 104 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.325 |
-0.022 |
| 105 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.320 |
-0.168 |
| 106 |
255907_at |
AT1G17920
|
HDG12, homeodomain GLABROUS 12 |
0.319 |
0.101 |
| 107 |
263233_at |
AT1G05577
|
unknown |
0.312 |
0.013 |
| 108 |
251636_at |
AT3G57530
|
ATCPK32, CDPK32, CPK32, calcium-dependent protein kinase 32 |
0.309 |
-0.008 |
| 109 |
267084_at |
AT2G41180
|
SIB2, sigma factor binding protein 2 |
0.308 |
-0.309 |
| 110 |
260032_at |
AT1G68750
|
ATPPC4, phosphoenolpyruvate carboxylase 4, PPC4, phosphoenolpyruvate carboxylase 4 |
0.306 |
-0.010 |
| 111 |
258985_at |
AT3G08960
|
[ARM repeat superfamily protein] |
0.305 |
0.108 |
| 112 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.302 |
0.031 |
| 113 |
253464_at |
AT4G32030
|
unknown |
0.299 |
0.093 |
| 114 |
265481_at |
AT2G15960
|
unknown |
0.298 |
0.072 |
| 115 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.297 |
-0.075 |
| 116 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
0.296 |
-0.147 |
| 117 |
265731_at |
AT2G01130
|
[DEA(D/H)-box RNA helicase family protein] |
0.295 |
0.022 |
| 118 |
253305_at |
AT4G33666
|
unknown |
0.288 |
0.027 |
| 119 |
267293_at |
AT2G23810
|
TET8, tetraspanin8 |
0.284 |
0.145 |
| 120 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.283 |
0.185 |
| 121 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.280 |
0.302 |
| 122 |
257441_at |
AT2G04020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.277 |
0.050 |
| 123 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.276 |
0.238 |
| 124 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.276 |
0.145 |
| 125 |
253322_at |
AT4G33980
|
unknown |
0.275 |
0.559 |
| 126 |
263163_at |
AT1G03160
|
FZL, FZO-like |
0.275 |
-0.096 |
| 127 |
257737_at |
AT3G27440
|
UKL5, uridine kinase-like 5 |
0.273 |
0.110 |
| 128 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
0.273 |
0.140 |
| 129 |
252930_at |
AT4G39010
|
AtGH9B18, glycosyl hydrolase 9B18, GH9B18, glycosyl hydrolase 9B18 |
0.273 |
0.088 |
| 130 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
0.272 |
0.194 |
| 131 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.268 |
0.027 |
| 132 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
0.267 |
0.090 |
| 133 |
253953_at |
AT4G26750
|
EXT-like, extensin-like, LIP5, homolog of mammalian LYST-INTERACTING PROTEIN5 |
0.266 |
0.211 |
| 134 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
0.266 |
-0.309 |
| 135 |
252207_at |
AT3G50370
|
unknown |
0.266 |
0.037 |
| 136 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
0.264 |
0.027 |
| 137 |
266053_at |
AT2G40730
|
CTEXP, cytoplasmic tRNA export protein |
0.263 |
0.151 |
| 138 |
250288_at |
AT5G13350
|
[Auxin-responsive GH3 family protein] |
0.263 |
0.070 |
| 139 |
250935_at |
AT5G03240
|
UBQ3, polyubiquitin 3 |
0.260 |
0.230 |
| 140 |
248981_at |
AT5G45110
|
ATNPR3, NPR3, NPR1-like protein 3 |
0.257 |
0.157 |
| 141 |
258382_at |
AT3G15355
|
PFU1, PHO2 FAMILY UBIQUITIN CONJUGATION ENZYME 1, UBC25, ubiquitin-conjugating enzyme 25 |
0.257 |
0.241 |
| 142 |
267449_at |
AT2G33690
|
[Late embryogenesis abundant protein, group 6] |
0.257 |
-0.079 |
| 143 |
261083_at |
AT1G07310
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.252 |
0.146 |
| 144 |
260477_at |
AT1G11050
|
[Protein kinase superfamily protein] |
0.252 |
0.100 |
| 145 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.252 |
0.125 |
| 146 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.252 |
0.075 |
| 147 |
260227_at |
AT1G74450
|
unknown |
0.249 |
0.053 |
| 148 |
249246_at |
AT5G42290
|
[transcription activator-related] |
0.248 |
0.056 |
| 149 |
258468_at |
AT3G06070
|
unknown |
0.247 |
-0.141 |
| 150 |
260576_at |
AT2G47310
|
[flowering time control protein-related / FCA gamma-related] |
0.246 |
0.017 |
| 151 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
0.246 |
-0.095 |
| 152 |
267381_at |
AT2G26190
|
[calmodulin-binding family protein] |
0.246 |
0.013 |
| 153 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
0.244 |
0.148 |
| 154 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.243 |
0.072 |
| 155 |
259012_at |
AT3G07360
|
ATPUB9, ARABIDOPSIS THALIANA PLANT U-BOX 9, PUB9, plant U-box 9 |
0.240 |
-0.124 |
| 156 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
0.239 |
0.116 |
| 157 |
265215_at |
AT1G05040
|
unknown |
0.239 |
0.041 |
| 158 |
246940_at |
AT5G25400
|
[Nucleotide-sugar transporter family protein] |
0.238 |
0.102 |
| 159 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
0.238 |
0.059 |
| 160 |
265552_at |
AT2G07560
|
AHA6, H(+)-ATPase 6, HA6, H(+)-ATPase 6 |
0.237 |
-0.004 |
| 161 |
246242_at |
AT4G36600
|
[Late embryogenesis abundant (LEA) protein] |
0.237 |
0.059 |
| 162 |
253808_at |
AT4G28300
|
unknown |
0.235 |
0.057 |
| 163 |
249748_at |
AT5G24620
|
[Pathogenesis-related thaumatin superfamily protein] |
0.235 |
0.236 |
| 164 |
250829_at |
AT5G04720
|
ADR1-L2, ADR1-like 2, PHX21, PHOENIX 21 |
0.234 |
0.019 |
| 165 |
255780_at |
AT1G18580
|
GAUT11, galacturonosyltransferase 11 |
0.234 |
0.161 |
| 166 |
261650_at |
AT1G27770
|
ACA1, autoinhibited Ca2+-ATPase 1, PEA1, PLASTID ENVELOPE ATPASE 1 |
0.232 |
0.096 |
| 167 |
247240_at |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
0.232 |
0.198 |
| 168 |
264880_at |
AT1G61210
|
DWA3, DWD hypersensitive to ABA 3 |
0.232 |
0.163 |
| 169 |
250321_at |
AT5G12850
|
[CCCH-type zinc finger protein with ARM repeat domain] |
0.231 |
0.077 |
| 170 |
245019_at |
ATCG00530
|
YCF10 |
0.230 |
-0.019 |
| 171 |
255815_at |
AT1G19890
|
ATMGH3, MALE-GAMETE-SPECIFIC HISTONE H3, MGH3, male-gamete-specific histone H3 |
0.230 |
0.093 |
| 172 |
262463_at |
AT1G50310
|
ATSTP9, SUGAR TRANSPORTER 9, STP9, sugar transporter 9 |
0.230 |
0.221 |
| 173 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.229 |
-0.095 |
| 174 |
260045_at |
AT1G73670
|
ATMPK15, MAP kinase 15, MPK15, MAP kinase 15 |
0.229 |
0.072 |
| 175 |
245906_at |
AT5G11070
|
unknown |
0.228 |
0.002 |
| 176 |
245901_at |
AT5G11060
|
KNAT4, KNOTTED1-like homeobox gene 4 |
0.228 |
0.074 |
| 177 |
265280_at |
AT2G28355
|
LCR5, low-molecular-weight cysteine-rich 5 |
0.226 |
0.078 |
| 178 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
0.226 |
-0.201 |
| 179 |
256955_at |
AT3G13390
|
sks11, SKU5 similar 11 |
0.226 |
0.065 |
| 180 |
246289_at |
AT3G56880
|
[VQ motif-containing protein] |
0.224 |
0.017 |
| 181 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.224 |
0.061 |
| 182 |
261499_at |
AT1G28430
|
CYP705A24, cytochrome P450, family 705, subfamily A, polypeptide 24 |
0.222 |
0.033 |
| 183 |
258990_at |
AT3G08840
|
[D-alanine--D-alanine ligase family] |
0.222 |
-0.147 |
| 184 |
257255_at |
AT3G21960
|
[Receptor-like protein kinase-related family protein] |
0.218 |
0.036 |
| 185 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.217 |
0.034 |
| 186 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.217 |
-0.011 |
| 187 |
244974_at |
ATCG00700
|
PSBN, photosystem II reaction center protein N |
-0.725 |
-0.020 |
| 188 |
253532_at |
AT4G31570
|
unknown |
-0.694 |
0.075 |
| 189 |
245008_at |
ATCG00360
|
YCF3 |
-0.547 |
-0.230 |
| 190 |
247719_at |
AT5G59305
|
unknown |
-0.426 |
0.040 |
| 191 |
256335_at |
AT1G72110
|
[O-acyltransferase (WSD1-like) family protein] |
-0.396 |
0.091 |
| 192 |
258289_at |
AT3G23450
|
unknown |
-0.375 |
-0.045 |
| 193 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
-0.371 |
-0.151 |
| 194 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.369 |
-0.283 |
| 195 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.368 |
0.026 |
| 196 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.362 |
-0.031 |
| 197 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
-0.339 |
0.083 |
| 198 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.336 |
-0.125 |
| 199 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.331 |
0.268 |
| 200 |
261764_at |
AT1G15530
|
LecRK-S.1, L-type lectin receptor kinase S.1 |
-0.328 |
0.072 |
| 201 |
253064_at |
AT4G37730
|
AtbZIP7, basic leucine-zipper 7, bZIP7, basic leucine-zipper 7 |
-0.327 |
0.000 |
| 202 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.327 |
-0.166 |
| 203 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.321 |
0.032 |
| 204 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.308 |
0.029 |
| 205 |
250372_at |
AT5G11460
|
unknown |
-0.307 |
0.063 |
| 206 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.295 |
-0.355 |
| 207 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.295 |
0.019 |
| 208 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.294 |
-0.065 |
| 209 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.277 |
0.025 |
| 210 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
-0.276 |
0.188 |
| 211 |
261345_at |
AT1G79760
|
DTA4, downstream target of AGL15-4 |
-0.272 |
0.061 |
| 212 |
264054_at |
AT2G22540
|
AGL22, AGAMOUS-like 22, FAQ1, Flowering Arabidopsis QTL1, SVP, SHORT VEGETATIVE PHASE |
-0.270 |
0.010 |
| 213 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.269 |
-0.064 |
| 214 |
250684_at |
AT5G06650
|
GIS2, GLABROUS INFLORESCENCE STEMS 2 |
-0.268 |
0.046 |
| 215 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.256 |
0.084 |
| 216 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.255 |
-0.094 |
| 217 |
245126_at |
AT2G47460
|
ATMYB12, MYB DOMAIN PROTEIN 12, MYB12, myb domain protein 12, PFG1, PRODUCTION OF FLAVONOL GLYCOSIDES 1 |
-0.252 |
0.042 |
| 218 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.247 |
0.073 |
| 219 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.247 |
0.068 |
| 220 |
263238_at |
AT2G16580
|
SAUR8, SMALL AUXIN UPREGULATED RNA 8 |
-0.243 |
0.070 |
| 221 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.242 |
-0.112 |
| 222 |
261840_at |
AT1G16070
|
AtTLP8, tubby like protein 8, TLP8, tubby like protein 8 |
-0.241 |
0.010 |
| 223 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
-0.240 |
-0.176 |
| 224 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.240 |
0.345 |
| 225 |
261861_at |
AT1G50450
|
[Saccharopine dehydrogenase ] |
-0.240 |
-0.417 |
| 226 |
259902_at |
AT1G74170
|
AtRLP13, receptor like protein 13, RLP13, receptor like protein 13 |
-0.237 |
-0.047 |
| 227 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.236 |
-0.297 |
| 228 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.236 |
-0.390 |
| 229 |
254145_at |
AT4G24700
|
unknown |
-0.234 |
-0.578 |
| 230 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.234 |
0.082 |
| 231 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
-0.233 |
-0.147 |
| 232 |
245800_at |
AT1G46264
|
AT-HSFB4, HSFB4, heat shock transcription factor B4, SCZ, SCHIZORIZA |
-0.233 |
0.117 |
| 233 |
262445_at |
AT1G47485
|
unknown |
-0.232 |
0.001 |
| 234 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.232 |
-0.054 |
| 235 |
259030_at |
AT3G09310
|
unknown |
-0.232 |
-0.031 |
| 236 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.230 |
0.036 |
| 237 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
-0.229 |
0.097 |
| 238 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
-0.228 |
-0.287 |
| 239 |
253689_at |
AT4G29770
|
unknown |
-0.225 |
0.145 |
| 240 |
251903_at |
AT3G54120
|
[Reticulon family protein] |
-0.225 |
0.092 |
| 241 |
248862_at |
AT5G46730
|
[glycine-rich protein] |
-0.225 |
0.070 |
| 242 |
257118_at |
AT3G20180
|
[Copper transport protein family] |
-0.224 |
0.036 |
| 243 |
262242_at |
AT1G48360
|
[zinc ion binding] |
-0.224 |
0.145 |
| 244 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
-0.223 |
0.039 |
| 245 |
253732_at |
AT4G29140
|
ADS1, ACTIVATED DISEASE SUSCEPTIBILITY 1 |
-0.222 |
-0.034 |
| 246 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
-0.220 |
0.226 |
| 247 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.220 |
0.106 |
| 248 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
-0.219 |
0.104 |
| 249 |
261447_at |
AT1G21160
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
-0.217 |
0.076 |
| 250 |
248572_at |
AT5G49800
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.216 |
0.025 |
| 251 |
257619_at |
AT3G24810
|
ICK3, KRP5, kip-related protein 5 |
-0.216 |
0.125 |
| 252 |
262338_at |
AT1G64185
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.214 |
-0.014 |
| 253 |
265376_at |
AT2G05810
|
[ARM repeat superfamily protein] |
-0.214 |
0.161 |
| 254 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.213 |
-0.001 |
| 255 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.211 |
-0.847 |
| 256 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
-0.210 |
0.015 |
| 257 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.210 |
0.463 |
| 258 |
264277_at |
AT1G60390
|
PG1, polygalacturonase 1 |
-0.209 |
0.088 |
| 259 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.209 |
0.197 |
| 260 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.208 |
-0.093 |
| 261 |
246797_at |
AT5G26790
|
unknown |
-0.208 |
0.138 |
| 262 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.205 |
-0.013 |
| 263 |
249029_at |
AT5G44870
|
LAZ5, LAZARUS 5, TTR1, tolerance to Tobacco ringspot virus 1 |
-0.205 |
0.015 |
| 264 |
254402_at |
AT4G21310
|
unknown |
-0.203 |
-0.210 |
| 265 |
253883_at |
AT4G27660
|
unknown |
-0.203 |
0.112 |
| 266 |
264331_at |
AT1G04130
|
AtTPR2, TPR2, tetratricopeptide repeat 2 |
-0.202 |
-0.097 |
| 267 |
250269_at |
AT5G12970
|
[Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase family protein] |
-0.201 |
0.088 |
| 268 |
258300_at |
AT3G23340
|
ckl10, casein kinase I-like 10 |
-0.198 |
-0.125 |
| 269 |
250719_at |
AT5G06250
|
DPA4, DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 |
-0.198 |
0.124 |
| 270 |
247319_at |
AT5G64050
|
ATERS, ERS, glutamate tRNA synthetase, OVA3, OVULE ABORTION 3 |
-0.198 |
0.105 |
| 271 |
256518_at |
AT1G66080
|
unknown |
-0.197 |
0.048 |
| 272 |
267636_at |
AT2G42110
|
unknown |
-0.197 |
0.042 |
| 273 |
260544_at |
AT2G43540
|
unknown |
-0.195 |
0.093 |
| 274 |
247966_at |
AT5G56610
|
[Phosphotyrosine protein phosphatases superfamily protein] |
-0.195 |
0.152 |
| 275 |
260677_at |
AT1G07910
|
AtRLG1, AtRLG1, ATRNL, ARABIDOPSIS THALIANA RNA LIGASE, RNL, RNAligase |
-0.194 |
-0.153 |
| 276 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.194 |
-0.099 |
| 277 |
261807_at |
AT1G30515
|
unknown |
-0.193 |
0.100 |
| 278 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.193 |
-0.228 |
| 279 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
-0.191 |
-0.002 |
| 280 |
252046_at |
AT3G52460
|
[hydroxyproline-rich glycoprotein family protein] |
-0.190 |
0.071 |
| 281 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.190 |
0.112 |
| 282 |
251477_at |
AT3G59680
|
unknown |
-0.189 |
0.072 |
| 283 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.189 |
-0.331 |
| 284 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
-0.188 |
0.272 |
| 285 |
250985_at |
AT5G02830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.188 |
-0.341 |
| 286 |
251271_at |
AT3G62050
|
[Putative endonuclease or glycosyl hydrolase] |
-0.186 |
0.006 |
| 287 |
255933_at |
AT1G12750
|
ATRBL6, RHOMBOID-like protein 6, RBL6, RHOMBOID-like protein 6 |
-0.185 |
0.038 |
| 288 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.185 |
-0.514 |
| 289 |
257277_at |
AT3G14470
|
[NB-ARC domain-containing disease resistance protein] |
-0.184 |
0.287 |
| 290 |
264049_at |
AT2G22390
|
ATRABA4E, RABA4E, RAB GTPase homolog A4E |
-0.184 |
0.052 |
| 291 |
261883_at |
AT1G80870
|
[Protein kinase superfamily protein] |
-0.184 |
0.101 |
| 292 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
-0.183 |
-0.051 |
| 293 |
252025_at |
AT3G52900
|
unknown |
-0.182 |
-0.024 |
| 294 |
255701_at |
AT4G00220
|
JLO, JAGGED LATERAL ORGANS, LBD30, LOB DOMAIN-CONTAINING PROTEIN 30 |
-0.181 |
0.074 |
| 295 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.181 |
-0.215 |
| 296 |
257646_at |
AT3G25740
|
MAP1B, methionine aminopeptidase 1C, MAP1C, METHIONINE AMINOPEPTIDASE 1C |
-0.180 |
-0.218 |
| 297 |
246019_at |
AT5G10690
|
[pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein] |
-0.180 |
-0.360 |
| 298 |
245194_at |
AT1G67820
|
[Protein phosphatase 2C family protein] |
-0.179 |
0.051 |
| 299 |
257648_at |
AT3G16840
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.179 |
0.002 |
| 300 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.179 |
-0.155 |
| 301 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
-0.178 |
-0.003 |
| 302 |
263805_at |
AT2G40400
|
unknown |
-0.177 |
-0.013 |
| 303 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
-0.177 |
0.040 |
| 304 |
248231_at |
AT5G53770
|
[Nucleotidyltransferase family protein] |
-0.177 |
0.024 |
| 305 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.176 |
-0.004 |
| 306 |
256784_at |
AT3G13674
|
unknown |
-0.176 |
0.047 |
| 307 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-0.176 |
0.201 |
| 308 |
259031_at |
AT3G09360
|
[Cyclin/Brf1-like TBP-binding protein] |
-0.175 |
-0.000 |
| 309 |
248438_at |
AT5G51230
|
AtEMF2, CYR1, EMF2, EMBRYONIC FLOWER 2, VEF2, CYTOKININ RESISTANT 1 |
-0.174 |
0.016 |
| 310 |
251734_at |
AT3G56270
|
unknown |
-0.174 |
-0.023 |
| 311 |
248097_at |
AT5G55260
|
PPX-2, PROTEIN PHOSPHATASE X -2, PPX2, protein phosphatase X 2 |
-0.174 |
-0.034 |
| 312 |
250201_at |
AT5G14230
|
unknown |
-0.174 |
0.171 |
| 313 |
262974_at |
AT1G75550
|
[glycine-rich protein] |
-0.173 |
0.064 |
| 314 |
263155_at |
AT1G54140
|
TAF9, TBP-ASSOCIATED FACTOR 9, TAFII21, TATA binding protein associated factor 21kDa subunit |
-0.173 |
0.086 |
| 315 |
252106_at |
AT3G51480
|
ATGLR3.6, glutamate receptor 3.6, GLR3.6, GLR3.6, glutamate receptor 3.6 |
-0.173 |
0.036 |
| 316 |
261910_at |
AT1G80730
|
ATZFP1, ARABIDOPSIS THALIANA ZINC-FINGER PROTEIN 1, ZFP1, zinc-finger protein 1 |
-0.172 |
0.041 |
| 317 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.172 |
-0.298 |
| 318 |
267219_at |
AT2G02590
|
unknown |
-0.172 |
0.024 |
| 319 |
257105_at |
AT3G15300
|
[VQ motif-containing protein] |
-0.171 |
0.201 |
| 320 |
247518_at |
AT5G61800
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.171 |
-0.003 |
| 321 |
257007_at |
AT3G14205
|
[Phosphoinositide phosphatase family protein] |
-0.170 |
-0.005 |
| 322 |
259489_at |
AT1G15790
|
unknown |
-0.170 |
0.331 |
| 323 |
251463_at |
AT3G59800
|
unknown |
-0.170 |
0.083 |
| 324 |
256049_at |
AT1G07010
|
AtSLP1, SLP1, Shewenella-like protein phosphatase 1 |
-0.170 |
-0.181 |
| 325 |
262620_at |
AT1G06540
|
unknown |
-0.169 |
0.064 |
| 326 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.169 |
0.087 |
| 327 |
254029_at |
AT4G25870
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.169 |
-0.085 |
| 328 |
262398_at |
AT1G49350
|
[pfkB-like carbohydrate kinase family protein] |
-0.169 |
-0.049 |
| 329 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.168 |
0.036 |
| 330 |
262388_at |
AT1G49320
|
unknown |
-0.168 |
-0.298 |
| 331 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.167 |
-0.025 |
| 332 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.166 |
-0.214 |
| 333 |
257852_at |
AT3G12950
|
[Trypsin family protein] |
-0.166 |
-0.052 |
| 334 |
261321_at |
AT1G44740
|
unknown |
-0.165 |
0.092 |
| 335 |
245078_at |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
-0.165 |
0.137 |
| 336 |
261928_at |
AT1G22480
|
[Cupredoxin superfamily protein] |
-0.165 |
0.113 |
| 337 |
248111_at |
AT5G55330
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.165 |
0.023 |
| 338 |
247234_at |
AT5G64980
|
unknown |
-0.164 |
0.100 |
| 339 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
-0.164 |
0.053 |
| 340 |
261587_at |
AT1G01660
|
[RING/U-box superfamily protein] |
-0.164 |
0.023 |
| 341 |
257191_at |
AT3G13175
|
unknown |
-0.163 |
0.154 |
| 342 |
253592_at |
AT4G30840
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.163 |
0.200 |
| 343 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.162 |
0.086 |
| 344 |
257665_at |
AT3G20430
|
unknown |
-0.162 |
-0.052 |
| 345 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
-0.162 |
-0.071 |
| 346 |
263151_at |
AT1G54120
|
unknown |
-0.162 |
-0.168 |
| 347 |
263712_at |
AT2G20585
|
NFD6, NUCLEAR FUSION DEFECTIVE 6 |
-0.162 |
0.022 |
| 348 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.162 |
-0.319 |
| 349 |
245943_at |
AT5G19500
|
[Tryptophan/tyrosine permease] |
-0.162 |
-0.117 |
| 350 |
254217_at |
AT4G23720
|
unknown |
-0.161 |
0.015 |
| 351 |
261591_at |
AT1G01740
|
BSK4, brassinosteroid-signaling kinase 4 |
-0.161 |
-0.100 |
| 352 |
246008_at |
AT5G08320
|
unknown |
-0.160 |
0.020 |
| 353 |
245818_at |
AT1G26100
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.159 |
0.138 |
| 354 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.159 |
0.037 |
| 355 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.158 |
-0.138 |
| 356 |
260685_at |
AT1G17650
|
AtGLYR2, GLYR2, glyoxylate reductase 2, GR2, GLYOXYLATE REDUCTASE 2 |
-0.158 |
-0.378 |
| 357 |
250694_at |
AT5G06710
|
HAT14, homeobox from Arabidopsis thaliana |
-0.158 |
0.025 |
| 358 |
251057_at |
AT5G01780
|
[2-oxoglutarate-dependent dioxygenase family protein] |
-0.157 |
-0.117 |
| 359 |
256334_at |
AT1G72125
|
AtNPF5.13, NPF5.13, NRT1/ PTR family 5.13 |
-0.156 |
0.105 |
| 360 |
266065_at |
AT2G18790
|
HY3, OOP1, OUT OF PHASE 1, PHYB, phytochrome B |
-0.156 |
-0.096 |
| 361 |
265742_at |
AT2G01290
|
RPI2, ribose-5-phosphate isomerase 2 |
-0.156 |
-0.174 |
| 362 |
255372_at |
AT4G04120
|
[copia-like retrotransposon family, has a 3.8e-19 P-value blast match to GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)gi|4996361|dbj|BAA78423.1| polyprotein (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.154 |
0.120 |
| 363 |
249189_at |
AT5G42780
|
AtHB27, homeobox protein 27, HB27, homeobox protein 27, ZHD13, ZINC FINGER HOMEODOMAIN 13 |
-0.154 |
0.088 |
| 364 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
-0.153 |
-0.184 |
| 365 |
249028_at |
AT5G44740
|
POLH, Y-family DNA polymerase H |
-0.153 |
-0.060 |
| 366 |
259299_at |
AT3G05080
|
unknown |
-0.152 |
-0.037 |
| 367 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.152 |
-0.814 |
| 368 |
267027_at |
AT2G38330
|
[MATE efflux family protein] |
-0.152 |
-0.108 |
| 369 |
264222_at |
AT1G60230
|
[Radical SAM superfamily protein] |
-0.152 |
-0.133 |
| 370 |
249762_at |
AT5G24000
|
unknown |
-0.152 |
-0.044 |
| 371 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
-0.152 |
-0.348 |