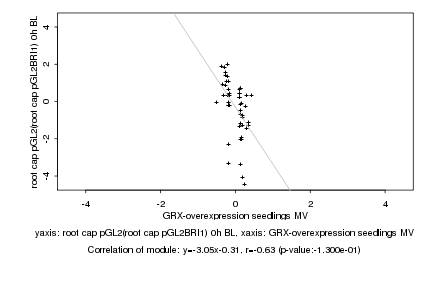
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
GRX-overexpression seedlings MV |
Log2 signal ratio
root cap pGL2(root cap pGL2BRI1) 0h BL |
| 1 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.413 |
0.339 |
| 2 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.349 |
-1.100 |
| 3 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
0.341 |
-1.259 |
| 4 |
250327_at |
AT5G12050
|
unknown |
0.284 |
-1.418 |
| 5 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.272 |
0.339 |
| 6 |
262349_at |
AT2G48130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.256 |
-0.258 |
| 7 |
261491_at |
AT1G14350
|
AtMYB124, myb domain protein 124, FLP, FOUR LIPS, MYB124 |
0.228 |
-4.402 |
| 8 |
246262_at |
AT1G31790
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.178 |
-0.704 |
| 9 |
263282_at |
AT2G14095
|
unknown |
0.177 |
-1.252 |
| 10 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
0.169 |
-4.048 |
| 11 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
0.162 |
-0.824 |
| 12 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
0.144 |
-2.026 |
| 13 |
248286_at |
AT5G52870
|
MAKR5, MEMBRANE-ASSOCIATED KINASE REGULATOR 5 |
0.137 |
-1.880 |
| 14 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.137 |
-0.080 |
| 15 |
254629_at |
AT4G18425
|
unknown |
0.131 |
-3.339 |
| 16 |
260074_at |
AT1G73640
|
AtRABA6a, RAB GTPase homolog A6A, RABA6a, RAB GTPase homolog A6A |
0.124 |
-0.479 |
| 17 |
248931_at |
AT5G46040
|
[Major facilitator superfamily protein] |
0.120 |
0.749 |
| 18 |
254536_at |
AT4G19720
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
0.119 |
-2.025 |
| 19 |
247992_at |
AT5G56520
|
unknown |
0.116 |
0.713 |
| 20 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
0.114 |
-1.139 |
| 21 |
265103_at |
AT1G31070
|
GlcNAc1pUT1, N-acetylglucosamine-1-phosphate uridylyltransferase 1 |
0.114 |
-0.685 |
| 22 |
246010_at |
AT5G08440
|
unknown |
0.113 |
-0.151 |
| 23 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
0.106 |
0.691 |
| 24 |
249787_at |
AT5G24320
|
[Transducin/WD40 repeat-like superfamily protein] |
0.097 |
0.234 |
| 25 |
265325_at |
AT2G18240
|
[Rer1 family protein] |
0.097 |
0.484 |
| 26 |
261277_at |
AT1G20230
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.097 |
-1.300 |
| 27 |
246625_at |
AT1G48880
|
TBL7, TRICHOME BIREFRINGENCE-LIKE 7 |
0.092 |
0.231 |
| 28 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.091 |
0.435 |
| 29 |
260423_at |
AT1G72470
|
ATEXO70D1, exocyst subunit exo70 family protein D1, EXO70D1, exocyst subunit exo70 family protein D1 |
0.090 |
0.237 |
| 30 |
244965_at |
ATCG00590
|
ORF31 |
-0.532 |
-0.032 |
| 31 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
-0.397 |
1.906 |
| 32 |
245025_at |
ATCG00130
|
ATPF |
-0.371 |
0.958 |
| 33 |
249669_at |
AT5G35880
|
unknown |
-0.334 |
0.339 |
| 34 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.304 |
1.827 |
| 35 |
249559_at |
AT5G38320
|
unknown |
-0.275 |
1.430 |
| 36 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
-0.270 |
1.573 |
| 37 |
257323_at |
ATMG01200
|
ORF294 |
-0.269 |
0.876 |
| 38 |
245026_at |
ATCG00140
|
ATPH |
-0.248 |
1.086 |
| 39 |
247560_at |
AT5G61090
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.235 |
0.339 |
| 40 |
245004_at |
ATCG00300
|
YCF9 |
-0.231 |
1.376 |
| 41 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.222 |
2.040 |
| 42 |
256840_x_at |
AT3G32000
|
[gypsy-like retrotransposon family (Athila), has a 5.4e-305 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.207 |
0.339 |
| 43 |
253356_at |
AT4G33390
|
unknown |
-0.201 |
0.339 |
| 44 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
-0.197 |
0.677 |
| 45 |
261987_at |
AT1G33710
|
[RNA-directed DNA polymerase (reverse transcriptase)-related family protein] |
-0.196 |
0.339 |
| 46 |
245785_at |
AT1G32180
|
ATCSLD6, cellulose synthase-like D6, CSLD6, CELLULOSE SYNTHASE LIKE D6, CSLD6, cellulose synthase-like D6 |
-0.194 |
-0.185 |
| 47 |
246363_at |
AT1G40390
|
[DNAse I-like superfamily protein] |
-0.194 |
0.339 |
| 48 |
251326_at |
AT3G61590
|
HS, HWS, HAWAIIAN SKIRT |
-0.194 |
-2.286 |
| 49 |
246857_at |
AT5G25920
|
unknown |
-0.194 |
-0.185 |
| 50 |
245020_at |
ATCG00540
|
PETA, photosynthetic electron transfer A |
-0.191 |
1.078 |
| 51 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
-0.191 |
-3.275 |
| 52 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.190 |
0.339 |
| 53 |
247017_at |
AT5G66980
|
[AP2/B3-like transcriptional factor family protein] |
-0.189 |
0.339 |
| 54 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.189 |
-0.010 |
| 55 |
258438_at |
AT3G17230
|
[invertase/pectin methylesterase inhibitor family protein] |
-0.180 |
-0.185 |
| 56 |
244935_at |
ATCG01090
|
NDHI |
-0.178 |
0.439 |
| 57 |
263723_at |
AT2G13500
|
unknown |
-0.176 |
0.339 |