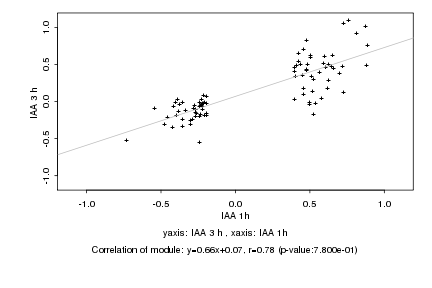
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
IAA 1h |
Log2 signal ratio
IAA 3 h |
| 1 |
246798_at |
AT5G26930
|
GATA23, GATA transcription factor 23 |
0.885 |
0.768 |
| 2 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
0.876 |
0.487 |
| 3 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.872 |
1.019 |
| 4 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
0.810 |
0.918 |
| 5 |
266611_at |
AT2G14960
|
GH3.1 |
0.756 |
1.106 |
| 6 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.725 |
1.062 |
| 7 |
253180_at |
AT4G35210
|
unknown |
0.725 |
0.126 |
| 8 |
257386_at |
AT2G42440
|
ASL15, ASYMMETRIC LEAVES 2-like 15, LBD17, LOB domain-containing protein 17 |
0.715 |
0.474 |
| 9 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.696 |
0.378 |
| 10 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.655 |
0.457 |
| 11 |
252103_at |
AT3G51410
|
unknown |
0.648 |
0.634 |
| 12 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
0.639 |
0.483 |
| 13 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.625 |
0.505 |
| 14 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.621 |
0.288 |
| 15 |
248789_at |
AT5G47440
|
unknown |
0.612 |
0.176 |
| 16 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.603 |
0.460 |
| 17 |
254459_at |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
0.592 |
0.608 |
| 18 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.586 |
0.520 |
| 19 |
252279_at |
AT3G49700
|
ACS9, 1-aminocyclopropane-1-carboxylate synthase 9, AtACS9, ETO3, ETHYLENE OVERPRODUCING 3 |
0.578 |
0.042 |
| 20 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.558 |
0.394 |
| 21 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.536 |
-0.021 |
| 22 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.522 |
-0.175 |
| 23 |
249992_at |
AT5G18560
|
PUCHI |
0.521 |
0.301 |
| 24 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
0.516 |
0.139 |
| 25 |
259027_at |
AT3G09280
|
unknown |
0.505 |
0.342 |
| 26 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.500 |
0.596 |
| 27 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
0.499 |
0.632 |
| 28 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
0.495 |
-0.035 |
| 29 |
247622_at |
AT5G60350
|
unknown |
0.492 |
-0.005 |
| 30 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.478 |
0.510 |
| 31 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.477 |
0.433 |
| 32 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.476 |
0.836 |
| 33 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.474 |
0.422 |
| 34 |
259735_at |
AT1G64405
|
unknown |
0.455 |
0.704 |
| 35 |
259223_at |
AT3G03660
|
WOX11, WUSCHEL related homeobox 11 |
0.454 |
0.105 |
| 36 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
0.454 |
0.185 |
| 37 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.449 |
0.353 |
| 38 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.431 |
0.502 |
| 39 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.423 |
0.543 |
| 40 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.420 |
0.659 |
| 41 |
245416_at |
AT4G17350
|
unknown |
0.408 |
0.499 |
| 42 |
253179_at |
AT4G35200
|
unknown |
0.402 |
0.338 |
| 43 |
266637_at |
AT2G35600
|
ATBRXL1, ARABIDOPSIS THALIANA BREVIS RADIX LIKE 1, BRXL1, BREVIS RADIX-like 1 |
0.396 |
0.029 |
| 44 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.394 |
0.465 |
| 45 |
247474_at |
AT5G62280
|
unknown |
0.392 |
0.407 |
| 46 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.737 |
-0.520 |
| 47 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
-0.546 |
-0.088 |
| 48 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.481 |
-0.307 |
| 49 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.459 |
-0.204 |
| 50 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.429 |
-0.350 |
| 51 |
256515_at |
AT1G66020
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.420 |
-0.055 |
| 52 |
266900_at |
AT2G34610
|
unknown |
-0.409 |
-0.008 |
| 53 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.396 |
-0.180 |
| 54 |
251916_at |
AT3G53960
|
[Major facilitator superfamily protein] |
-0.394 |
0.034 |
| 55 |
256788_at |
AT3G13730
|
CYP90D1, cytochrome P450, family 90, subfamily D, polypeptide 1 |
-0.385 |
-0.124 |
| 56 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
-0.382 |
-0.030 |
| 57 |
262811_at |
AT1G11700
|
unknown |
-0.361 |
-0.229 |
| 58 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
-0.358 |
0.000 |
| 59 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.357 |
-0.325 |
| 60 |
246900_at |
AT5G25620
|
AtYUC6, YUC6, YUCCA6 |
-0.340 |
-0.114 |
| 61 |
250872_at |
AT5G03960
|
IQD12, IQ-domain 12 |
-0.308 |
-0.247 |
| 62 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.302 |
-0.304 |
| 63 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.292 |
-0.231 |
| 64 |
264305_at |
AT1G78815
|
LSH7, LIGHT SENSITIVE HYPOCOTYLS 7 |
-0.285 |
-0.081 |
| 65 |
252626_at |
AT3G44940
|
unknown |
-0.281 |
-0.050 |
| 66 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.273 |
-0.195 |
| 67 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.273 |
-0.144 |
| 68 |
248140_at |
AT5G54980
|
[Uncharacterised protein family (UPF0497)] |
-0.270 |
-0.099 |
| 69 |
251058_at |
AT5G01790
|
unknown |
-0.262 |
-0.160 |
| 70 |
266403_at |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.245 |
-0.061 |
| 71 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
-0.243 |
-0.197 |
| 72 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.242 |
-0.546 |
| 73 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.242 |
-0.002 |
| 74 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
-0.238 |
-0.167 |
| 75 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
-0.236 |
-0.041 |
| 76 |
261545_at |
AT1G63530
|
unknown |
-0.232 |
0.034 |
| 77 |
254240_at |
AT4G23496
|
SP1L5, SPIRAL1-like5 |
-0.230 |
-0.032 |
| 78 |
261441_at |
AT1G28470
|
ANAC010, NAC domain containing protein 10, NAC010, NAC domain containing protein 10, SND3, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 |
-0.228 |
-0.042 |
| 79 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
-0.222 |
-0.100 |
| 80 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.222 |
-0.021 |
| 81 |
265014_at |
AT1G24430
|
[HXXXD-type acyl-transferase family protein] |
-0.222 |
-0.067 |
| 82 |
249042_at |
AT5G44350
|
[ethylene-responsive nuclear protein -related] |
-0.219 |
-0.018 |
| 83 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.217 |
0.092 |
| 84 |
259857_at |
AT1G68430
|
unknown |
-0.210 |
-0.010 |
| 85 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.208 |
-0.185 |
| 86 |
256154_at |
AT3G08490
|
unknown |
-0.201 |
-0.156 |
| 87 |
254434_at |
AT4G20880
|
[ethylene-responsive nuclear protein / ethylene-regulated nuclear protein (ERT2)] |
-0.201 |
-0.019 |
| 88 |
263915_at |
AT2G36430
|
unknown |
-0.199 |
-0.188 |
| 89 |
247470_at |
AT5G62220
|
ATGT18, glycosyltransferase 18, GT18, glycosyltransferase 18, XLT2, Xyloglucan L-side chain galactosylTransferase position 2 |
-0.196 |
0.069 |