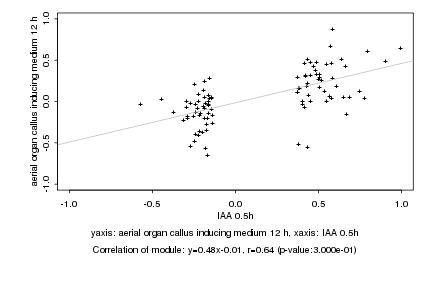
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
IAA 0.5h |
Log2 signal ratio
aerial organ callus inducing medium 12 h |
| 1 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
0.992 |
0.650 |
| 2 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.899 |
0.486 |
| 3 |
266611_at |
AT2G14960
|
GH3.1 |
0.791 |
0.609 |
| 4 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
0.774 |
0.044 |
| 5 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.742 |
0.125 |
| 6 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.682 |
0.057 |
| 7 |
248789_at |
AT5G47440
|
unknown |
0.667 |
-0.145 |
| 8 |
257386_at |
AT2G42440
|
ASL15, ASYMMETRIC LEAVES 2-like 15, LBD17, LOB domain-containing protein 17 |
0.661 |
0.428 |
| 9 |
259027_at |
AT3G09280
|
unknown |
0.648 |
0.060 |
| 10 |
246798_at |
AT5G26930
|
GATA23, GATA transcription factor 23 |
0.635 |
0.519 |
| 11 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.603 |
0.185 |
| 12 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
0.581 |
0.883 |
| 13 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.581 |
0.288 |
| 14 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
0.578 |
0.464 |
| 15 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.573 |
0.038 |
| 16 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.567 |
0.668 |
| 17 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
0.564 |
0.072 |
| 18 |
247622_at |
AT5G60350
|
unknown |
0.547 |
0.006 |
| 19 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.547 |
0.454 |
| 20 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.535 |
0.129 |
| 21 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.515 |
0.260 |
| 22 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.505 |
0.181 |
| 23 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.505 |
0.302 |
| 24 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.502 |
0.336 |
| 25 |
252765_at |
AT3G42800
|
unknown |
0.500 |
0.278 |
| 26 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.487 |
0.331 |
| 27 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.486 |
0.479 |
| 28 |
252103_at |
AT3G51410
|
unknown |
0.481 |
0.380 |
| 29 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
0.468 |
0.429 |
| 30 |
253047_at |
AT4G37295
|
unknown |
0.449 |
0.484 |
| 31 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.449 |
0.325 |
| 32 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
0.448 |
0.005 |
| 33 |
253910_at |
AT4G27290
|
[S-locus lectin protein kinase family protein] |
0.435 |
0.079 |
| 34 |
245140_at |
AT2G45420
|
LBD18, LOB domain-containing protein 18 |
0.433 |
0.515 |
| 35 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
0.431 |
-0.549 |
| 36 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
0.429 |
0.225 |
| 37 |
249992_at |
AT5G18560
|
PUCHI |
0.428 |
0.184 |
| 38 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.421 |
0.313 |
| 39 |
251751_at |
AT3G55720
|
unknown |
0.418 |
0.317 |
| 40 |
251940_at |
AT3G53450
|
LOG4, LONELY GUY 4 |
0.414 |
-0.066 |
| 41 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.413 |
0.467 |
| 42 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.400 |
-0.027 |
| 43 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.398 |
0.007 |
| 44 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.383 |
0.158 |
| 45 |
248801_at |
AT5G47370
|
HAT2 |
0.379 |
0.165 |
| 46 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.378 |
-0.510 |
| 47 |
267250_at |
AT2G23060
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.373 |
0.116 |
| 48 |
247474_at |
AT5G62280
|
unknown |
0.369 |
0.292 |
| 49 |
266637_at |
AT2G35600
|
ATBRXL1, ARABIDOPSIS THALIANA BREVIS RADIX LIKE 1, BRXL1, BREVIS RADIX-like 1 |
0.368 |
0.113 |
| 50 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.577 |
-0.025 |
| 51 |
266900_at |
AT2G34610
|
unknown |
-0.449 |
0.028 |
| 52 |
256788_at |
AT3G13730
|
CYP90D1, cytochrome P450, family 90, subfamily D, polypeptide 1 |
-0.377 |
-0.128 |
| 53 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.314 |
-0.226 |
| 54 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
-0.299 |
-0.061 |
| 55 |
252236_at |
AT3G49930
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.297 |
0.010 |
| 56 |
264305_at |
AT1G78815
|
LSH7, LIGHT SENSITIVE HYPOCOTYLS 7 |
-0.294 |
-0.181 |
| 57 |
261073_at |
AT1G07300
|
[josephin protein-related] |
-0.292 |
-0.196 |
| 58 |
256066_at |
AT1G06980
|
unknown |
-0.276 |
-0.017 |
| 59 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.274 |
-0.542 |
| 60 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.258 |
-0.179 |
| 61 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.252 |
-0.476 |
| 62 |
250872_at |
AT5G03960
|
IQD12, IQ-domain 12 |
-0.252 |
0.213 |
| 63 |
246900_at |
AT5G25620
|
AtYUC6, YUC6, YUCCA6 |
-0.245 |
-0.126 |
| 64 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.244 |
-0.398 |
| 65 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.242 |
-0.033 |
| 66 |
245560_at |
AT4G15480
|
UGT84A1 |
-0.233 |
-0.078 |
| 67 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.225 |
0.009 |
| 68 |
256450_at |
AT1G75290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.225 |
0.090 |
| 69 |
246231_at |
AT4G37080
|
unknown |
-0.223 |
-0.404 |
| 70 |
262811_at |
AT1G11700
|
unknown |
-0.220 |
-0.358 |
| 71 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
-0.218 |
-0.161 |
| 72 |
250882_at |
AT5G04000
|
unknown |
-0.216 |
-0.138 |
| 73 |
260155_at |
AT1G52870
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.201 |
-0.374 |
| 74 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.195 |
0.145 |
| 75 |
254263_at |
AT4G23493
|
unknown |
-0.193 |
-0.052 |
| 76 |
250908_at |
AT5G03680
|
PTL, PETAL LOSS |
-0.192 |
-0.076 |
| 77 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.192 |
0.246 |
| 78 |
249184_at |
AT5G43020
|
[Leucine-rich repeat protein kinase family protein] |
-0.188 |
-0.200 |
| 79 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.188 |
0.057 |
| 80 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.186 |
-0.560 |
| 81 |
246969_at |
AT5G24880
|
unknown |
-0.185 |
-0.019 |
| 82 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.179 |
-0.340 |
| 83 |
264238_at |
AT1G54740
|
unknown |
-0.176 |
-0.276 |
| 84 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.169 |
-0.644 |
| 85 |
246580_at |
AT1G31770
|
ABCG14, ATP-binding cassette G14 |
-0.169 |
-0.201 |
| 86 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.168 |
0.076 |
| 87 |
266635_at |
AT2G35470
|
unknown |
-0.168 |
0.006 |
| 88 |
257483_at |
AT1G49620
|
AtKRP7, ICK5, ICN6, KRP7, KIP-RELATED PROTEIN 7 |
-0.166 |
-0.048 |
| 89 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.165 |
-0.139 |
| 90 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
-0.164 |
-0.031 |
| 91 |
246316_at |
AT3G56890
|
[F-box associated ubiquitination effector family protein] |
-0.161 |
0.037 |
| 92 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
-0.161 |
0.284 |
| 93 |
248004_at |
AT5G56230
|
PRA1.G2, prenylated RAB acceptor 1.G2 |
-0.150 |
0.046 |
| 94 |
252628_at |
AT3G44960
|
unknown |
-0.148 |
-0.093 |
| 95 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.146 |
0.050 |
| 96 |
247340_at |
AT5G63700
|
[zinc ion binding] |
-0.142 |
-0.162 |
| 97 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
-0.142 |
-0.265 |