|
probeID |
AGICode |
Annotation |
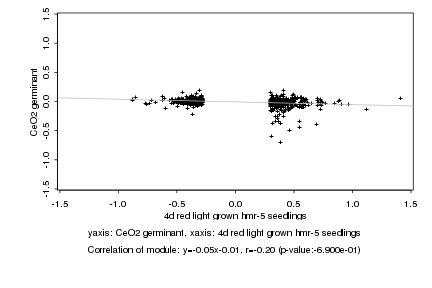
Log2 signal ratio
4d red light grown hmr-5 seedlings |
Log2 signal ratio
CeO2 germinant |
| 1 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
1.407 |
0.062 |
| 2 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
1.114 |
-0.134 |
| 3 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.960 |
-0.046 |
| 4 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.904 |
-0.041 |
| 5 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.888 |
0.020 |
| 6 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.873 |
0.015 |
| 7 |
245233_at |
AT4G25580
|
[CAP160 protein] |
0.839 |
-0.022 |
| 8 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.763 |
-0.031 |
| 9 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.742 |
-0.014 |
| 10 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.734 |
0.010 |
| 11 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.727 |
-0.061 |
| 12 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.725 |
-0.122 |
| 13 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.723 |
-0.047 |
| 14 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
0.721 |
0.045 |
| 15 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.706 |
-0.012 |
| 16 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.697 |
-0.057 |
| 17 |
253322_at |
AT4G33980
|
unknown |
0.695 |
0.024 |
| 18 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
0.695 |
0.055 |
| 19 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
0.688 |
-0.385 |
| 20 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.649 |
-0.030 |
| 21 |
266518_at |
AT2G35170
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.619 |
-0.053 |
| 22 |
244999_at |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
0.609 |
-0.003 |
| 23 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.606 |
0.046 |
| 24 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.596 |
0.036 |
| 25 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.595 |
-0.053 |
| 26 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.590 |
0.020 |
| 27 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.589 |
0.017 |
| 28 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.581 |
0.067 |
| 29 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.578 |
-0.044 |
| 30 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.577 |
-0.091 |
| 31 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.570 |
0.049 |
| 32 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.569 |
-0.066 |
| 33 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.569 |
0.034 |
| 34 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.567 |
-0.058 |
| 35 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.566 |
-0.086 |
| 36 |
260668_at |
AT1G19530
|
unknown |
0.565 |
-0.097 |
| 37 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.564 |
-0.069 |
| 38 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.563 |
-0.098 |
| 39 |
258327_at |
AT3G22640
|
PAP85 |
0.562 |
-0.092 |
| 40 |
263881_at |
AT2G21820
|
unknown |
0.562 |
0.005 |
| 41 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
0.561 |
0.078 |
| 42 |
259982_at |
AT1G76410
|
ATL8 |
0.557 |
0.080 |
| 43 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.552 |
0.086 |
| 44 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.551 |
-0.010 |
| 45 |
260744_at |
AT1G15010
|
unknown |
0.548 |
-0.012 |
| 46 |
248585_at |
AT5G49640
|
unknown |
0.548 |
-0.006 |
| 47 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.542 |
-0.332 |
| 48 |
255733_at |
AT1G25400
|
unknown |
0.542 |
-0.090 |
| 49 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
0.541 |
-0.039 |
| 50 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.540 |
-0.442 |
| 51 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.530 |
-0.084 |
| 52 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.527 |
0.056 |
| 53 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.527 |
0.073 |
| 54 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.522 |
0.064 |
| 55 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.512 |
-0.068 |
| 56 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
0.512 |
0.073 |
| 57 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.509 |
-0.043 |
| 58 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.507 |
0.035 |
| 59 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.506 |
0.038 |
| 60 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.501 |
-0.032 |
| 61 |
253802_at |
AT4G28180
|
unknown |
0.501 |
0.071 |
| 62 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.501 |
0.006 |
| 63 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.494 |
0.126 |
| 64 |
249454_at |
AT5G39520
|
unknown |
0.491 |
0.014 |
| 65 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.486 |
-0.010 |
| 66 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.485 |
0.070 |
| 67 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.484 |
0.051 |
| 68 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.483 |
-0.093 |
| 69 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.482 |
0.115 |
| 70 |
244938_at |
ATCG01120
|
RPS15, chloroplast ribosomal protein S15 |
0.482 |
-0.069 |
| 71 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
0.482 |
0.062 |
| 72 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
0.480 |
0.025 |
| 73 |
250770_at |
AT5G05390
|
LAC12, laccase 12 |
0.479 |
-0.027 |
| 74 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
0.478 |
-0.013 |
| 75 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.476 |
-0.057 |
| 76 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.467 |
-0.107 |
| 77 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
0.467 |
-0.013 |
| 78 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.466 |
0.047 |
| 79 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
0.465 |
0.006 |
| 80 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.462 |
-0.017 |
| 81 |
264774_at |
AT1G22890
|
unknown |
0.461 |
-0.129 |
| 82 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.455 |
-0.491 |
| 83 |
245209_at |
AT5G12340
|
unknown |
0.453 |
-0.140 |
| 84 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
0.452 |
0.044 |
| 85 |
263475_at |
AT2G31945
|
unknown |
0.451 |
-0.086 |
| 86 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.451 |
-0.032 |
| 87 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.451 |
0.033 |
| 88 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.449 |
-0.000 |
| 89 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.447 |
0.013 |
| 90 |
261205_at |
AT1G12790
|
unknown |
0.446 |
-0.104 |
| 91 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
0.444 |
-0.007 |
| 92 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.439 |
0.016 |
| 93 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.438 |
-0.002 |
| 94 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.438 |
-0.052 |
| 95 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.438 |
-0.014 |
| 96 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.438 |
-0.051 |
| 97 |
252539_at |
AT3G45730
|
unknown |
0.435 |
0.063 |
| 98 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.433 |
0.041 |
| 99 |
255285_at |
AT4G04630
|
unknown |
0.429 |
0.031 |
| 100 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.427 |
-0.059 |
| 101 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.426 |
-0.001 |
| 102 |
258383_at |
AT3G15440
|
unknown |
0.424 |
0.015 |
| 103 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.423 |
0.003 |
| 104 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.423 |
-0.126 |
| 105 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.421 |
0.012 |
| 106 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.421 |
0.039 |
| 107 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.419 |
-0.169 |
| 108 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
0.418 |
-0.015 |
| 109 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.417 |
-0.032 |
| 110 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.415 |
-0.016 |
| 111 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.414 |
0.030 |
| 112 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
0.412 |
0.028 |
| 113 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.411 |
-0.004 |
| 114 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.411 |
0.004 |
| 115 |
248432_at |
AT5G51390
|
unknown |
0.411 |
-0.001 |
| 116 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
0.410 |
-0.038 |
| 117 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.409 |
0.192 |
| 118 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.408 |
-0.182 |
| 119 |
247774_at |
AT5G58660
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.408 |
0.008 |
| 120 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.407 |
0.053 |
| 121 |
257670_at |
AT3G20340
|
unknown |
0.406 |
0.088 |
| 122 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.405 |
0.126 |
| 123 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
0.405 |
-0.052 |
| 124 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.404 |
-0.255 |
| 125 |
244997_at |
ATCG00170
|
RPOC2 |
0.403 |
-0.087 |
| 126 |
258263_at |
AT3G15780
|
unknown |
0.403 |
-0.038 |
| 127 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.402 |
0.028 |
| 128 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.399 |
-0.053 |
| 129 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.398 |
0.048 |
| 130 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.397 |
-0.054 |
| 131 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
0.397 |
-0.005 |
| 132 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.397 |
-0.076 |
| 133 |
265083_at |
AT1G03820
|
unknown |
0.394 |
0.032 |
| 134 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.394 |
-0.022 |
| 135 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.391 |
-0.060 |
| 136 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.391 |
-0.098 |
| 137 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
0.390 |
-0.007 |
| 138 |
256061_at |
AT1G07040
|
unknown |
0.389 |
0.016 |
| 139 |
261673_at |
AT1G18280
|
LTPG3, glycosylphosphatidylinositol-anchored lipid protein transfer 3 |
0.389 |
0.033 |
| 140 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.388 |
-0.149 |
| 141 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
0.388 |
0.028 |
| 142 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.388 |
-0.055 |
| 143 |
262359_at |
AT1G73066
|
[Leucine-rich repeat family protein] |
0.387 |
-0.058 |
| 144 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.387 |
0.116 |
| 145 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.384 |
-0.054 |
| 146 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.383 |
0.029 |
| 147 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.383 |
-0.127 |
| 148 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.383 |
-0.701 |
| 149 |
246584_at |
AT5G14730
|
unknown |
0.382 |
0.052 |
| 150 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.382 |
-0.033 |
| 151 |
266658_at |
AT2G25735
|
unknown |
0.380 |
-0.366 |
| 152 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
0.379 |
0.096 |
| 153 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.379 |
0.015 |
| 154 |
260656_at |
AT1G19380
|
unknown |
0.379 |
-0.079 |
| 155 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.378 |
0.010 |
| 156 |
264635_at |
AT1G65500
|
unknown |
0.378 |
0.055 |
| 157 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.376 |
-0.007 |
| 158 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.375 |
-0.021 |
| 159 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.374 |
-0.010 |
| 160 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.373 |
-0.213 |
| 161 |
264821_at |
AT1G03470
|
NET3A, Networked 3A |
0.372 |
0.041 |
| 162 |
267036_at |
AT2G38465
|
unknown |
0.371 |
-0.040 |
| 163 |
247004_at |
AT5G67570
|
DG1, DELAYED GREENING 1, EMB1408, embryo defective 1408, EMB246, EMBRYO DEFECTIVE 246 |
0.371 |
-0.020 |
| 164 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
0.370 |
-0.006 |
| 165 |
264362_at |
AT1G03290
|
unknown |
0.370 |
0.022 |
| 166 |
262607_at |
AT1G13990
|
unknown |
0.369 |
-0.021 |
| 167 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.368 |
-0.024 |
| 168 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
0.368 |
0.017 |
| 169 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.367 |
-0.080 |
| 170 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.365 |
-0.326 |
| 171 |
260536_at |
AT2G43400
|
ETFQO, electron-transfer flavoprotein:ubiquinone oxidoreductase |
0.365 |
-0.036 |
| 172 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.364 |
0.044 |
| 173 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.364 |
-0.098 |
| 174 |
250873_at |
AT5G03980
|
[SGNH hydrolase-type esterase superfamily protein] |
0.364 |
0.005 |
| 175 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.363 |
0.022 |
| 176 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.363 |
0.002 |
| 177 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.363 |
-0.053 |
| 178 |
244998_at |
ATCG00180
|
RPOC1 |
0.363 |
-0.064 |
| 179 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.362 |
0.012 |
| 180 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.362 |
-0.050 |
| 181 |
260272_at |
AT1G80570
|
[RNI-like superfamily protein] |
0.361 |
-0.095 |
| 182 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.361 |
0.033 |
| 183 |
263118_at |
AT1G03090
|
MCCA |
0.361 |
-0.009 |
| 184 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.358 |
-0.244 |
| 185 |
263113_at |
AT1G03150
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.356 |
-0.041 |
| 186 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
0.356 |
0.029 |
| 187 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.356 |
-0.290 |
| 188 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
0.355 |
0.005 |
| 189 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
0.354 |
-0.073 |
| 190 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
0.354 |
-0.043 |
| 191 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.354 |
-0.006 |
| 192 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.354 |
-0.169 |
| 193 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.354 |
-0.002 |
| 194 |
246963_at |
AT5G24820
|
[Eukaryotic aspartyl protease family protein] |
0.353 |
0.023 |
| 195 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.353 |
0.073 |
| 196 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.352 |
-0.031 |
| 197 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
0.351 |
0.021 |
| 198 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.351 |
0.034 |
| 199 |
252095_at |
AT3G51000
|
[alpha/beta-Hydrolases superfamily protein] |
0.351 |
-0.014 |
| 200 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.349 |
0.063 |
| 201 |
260930_at |
AT1G02620
|
[Ras-related small GTP-binding family protein] |
0.349 |
0.007 |
| 202 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.348 |
0.037 |
| 203 |
248713_at |
AT5G48180
|
AtNSP5, NSP5, nitrile specifier protein 5 |
0.348 |
0.046 |
| 204 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.348 |
0.048 |
| 205 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.346 |
0.077 |
| 206 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.346 |
0.036 |
| 207 |
251022_at |
AT5G02150
|
Fes1C, Fes1C |
0.346 |
-0.005 |
| 208 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
0.346 |
-0.060 |
| 209 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.346 |
-0.063 |
| 210 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.345 |
0.116 |
| 211 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.345 |
-0.015 |
| 212 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.344 |
-0.050 |
| 213 |
264405_at |
AT1G10210
|
ATMPK1, mitogen-activated protein kinase 1, MPK1, mitogen-activated protein kinase 1 |
0.343 |
0.027 |
| 214 |
257580_at |
AT3G06210
|
[ARM repeat superfamily protein] |
0.343 |
0.062 |
| 215 |
262322_at |
AT1G27590
|
unknown |
0.343 |
-0.074 |
| 216 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.343 |
-0.111 |
| 217 |
263281_at |
AT2G14160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.342 |
0.045 |
| 218 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
0.342 |
0.017 |
| 219 |
265989_at |
AT2G24260
|
LRL1, LJRHL1-like 1 |
0.342 |
-0.010 |
| 220 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
0.342 |
-0.012 |
| 221 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.341 |
-0.080 |
| 222 |
263265_at |
AT2G38820
|
unknown |
0.341 |
-0.016 |
| 223 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.341 |
-0.043 |
| 224 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.341 |
-0.326 |
| 225 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
0.341 |
0.066 |
| 226 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.341 |
-0.081 |
| 227 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
0.340 |
0.008 |
| 228 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.339 |
-0.251 |
| 229 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.338 |
0.019 |
| 230 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
0.337 |
0.042 |
| 231 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.337 |
-0.043 |
| 232 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.336 |
0.024 |
| 233 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
0.334 |
-0.048 |
| 234 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.332 |
-0.113 |
| 235 |
254445_at |
AT4G20930
|
[6-phosphogluconate dehydrogenase family protein] |
0.331 |
-0.052 |
| 236 |
264236_at |
AT1G54680
|
unknown |
0.330 |
-0.022 |
| 237 |
251166_at |
AT3G63350
|
AT-HSFA7B, HSFA7B, HEAT SHOCK TRANSCRIPTION FACTOR A7B |
0.330 |
0.050 |
| 238 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.329 |
0.049 |
| 239 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.328 |
-0.049 |
| 240 |
253126_at |
AT4G36050
|
[endonuclease/exonuclease/phosphatase family protein] |
0.327 |
-0.004 |
| 241 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.326 |
0.020 |
| 242 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.326 |
-0.038 |
| 243 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
0.325 |
-0.022 |
| 244 |
246929_at |
AT5G25210
|
unknown |
0.325 |
0.047 |
| 245 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.324 |
0.005 |
| 246 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.323 |
-0.141 |
| 247 |
247197_at |
AT5G65240
|
[Leucine-rich repeat protein kinase family protein] |
0.322 |
-0.067 |
| 248 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
0.322 |
0.036 |
| 249 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.320 |
0.084 |
| 250 |
259979_at |
AT1G76600
|
unknown |
0.319 |
-0.058 |
| 251 |
248205_at |
AT5G54300
|
unknown |
0.318 |
-0.035 |
| 252 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
0.318 |
0.011 |
| 253 |
245781_at |
AT1G45976
|
SBP1, S-ribonuclease binding protein 1 |
0.317 |
0.001 |
| 254 |
265266_at |
AT2G42890
|
AML2, MEI2-like 2, ML2, MEI2-like 2 |
0.317 |
0.005 |
| 255 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.316 |
-0.360 |
| 256 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.314 |
-0.017 |
| 257 |
249736_at |
AT5G24460
|
unknown |
0.313 |
-0.002 |
| 258 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.313 |
0.013 |
| 259 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.312 |
-0.061 |
| 260 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.312 |
-0.024 |
| 261 |
266910_at |
AT2G45920
|
[U-box domain-containing protein] |
0.312 |
-0.104 |
| 262 |
266753_at |
AT2G46990
|
IAA20, indole-3-acetic acid inducible 20 |
0.310 |
-0.007 |
| 263 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
0.310 |
-0.022 |
| 264 |
245314_at |
AT4G16745
|
[Exostosin family protein] |
0.310 |
0.037 |
| 265 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.310 |
0.108 |
| 266 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.310 |
0.073 |
| 267 |
247992_at |
AT5G56520
|
unknown |
0.310 |
0.017 |
| 268 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
0.309 |
-0.005 |
| 269 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.309 |
-0.036 |
| 270 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.308 |
0.039 |
| 271 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.307 |
-0.022 |
| 272 |
251291_at |
AT3G61900
|
SAUR33, SMALL AUXIN UPREGULATED RNA 33 |
0.307 |
0.023 |
| 273 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.307 |
0.058 |
| 274 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
0.306 |
-0.007 |
| 275 |
254200_at |
AT4G24110
|
unknown |
0.306 |
-0.159 |
| 276 |
264379_at |
AT2G25200
|
unknown |
0.306 |
-0.019 |
| 277 |
260732_at |
AT1G17520
|
[Homeodomain-like/winged-helix DNA-binding family protein] |
0.305 |
0.034 |
| 278 |
262977_at |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
0.304 |
-0.028 |
| 279 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.304 |
-0.056 |
| 280 |
247616_at |
AT5G60260
|
unknown |
0.303 |
-0.046 |
| 281 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.302 |
0.035 |
| 282 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
0.302 |
0.107 |
| 283 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
0.302 |
0.011 |
| 284 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.302 |
-0.154 |
| 285 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.301 |
-0.022 |
| 286 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.301 |
-0.038 |
| 287 |
252117_at |
AT3G51430
|
SSL5, STRICTOSIDINE SYNTHASE-LIKE 5, YLS2, YELLOW-LEAF-SPECIFIC GENE 2 |
0.300 |
-0.052 |
| 288 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.300 |
-0.598 |
| 289 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.299 |
-0.033 |
| 290 |
246254_at |
AT4G36450
|
ATMPK14, mitogen-activated protein kinase 14, MPK14, MPK14, mitogen-activated protein kinase 14 |
0.299 |
-0.062 |
| 291 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.299 |
-0.145 |
| 292 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.298 |
0.166 |
| 293 |
261558_at |
AT1G01770
|
unknown |
0.298 |
-0.073 |
| 294 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
0.298 |
0.050 |
| 295 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
0.298 |
-0.025 |
| 296 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.297 |
-0.010 |
| 297 |
260929_at |
AT1G02680
|
TAF13, TBP-associated factor 13 |
0.297 |
0.027 |
| 298 |
248879_at |
AT5G46180
|
DELTA-OAT, ornithine-delta-aminotransferase |
0.297 |
-0.038 |
| 299 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.296 |
0.015 |
| 300 |
253279_at |
AT4G34030
|
MCCB, 3-methylcrotonyl-CoA carboxylase |
0.294 |
-0.006 |
| 301 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.884 |
0.042 |
| 302 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.881 |
0.031 |
| 303 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.857 |
0.086 |
| 304 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.777 |
-0.022 |
| 305 |
244972_at |
ATCG00680
|
PSBB, photosystem II reaction center protein B |
-0.766 |
-0.042 |
| 306 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.741 |
-0.025 |
| 307 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.721 |
0.032 |
| 308 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
-0.688 |
-0.012 |
| 309 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.628 |
0.027 |
| 310 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.624 |
0.092 |
| 311 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.609 |
0.064 |
| 312 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.600 |
-0.118 |
| 313 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.571 |
0.030 |
| 314 |
250327_at |
AT5G12050
|
unknown |
-0.549 |
0.041 |
| 315 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.548 |
-0.031 |
| 316 |
251058_at |
AT5G01790
|
unknown |
-0.542 |
0.020 |
| 317 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.539 |
0.023 |
| 318 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.538 |
0.047 |
| 319 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.533 |
-0.025 |
| 320 |
248074_at |
AT5G55730
|
FLA1, FASCICLIN-like arabinogalactan 1 |
-0.523 |
0.006 |
| 321 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.522 |
-0.009 |
| 322 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.518 |
0.017 |
| 323 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
-0.515 |
0.005 |
| 324 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.515 |
0.032 |
| 325 |
261949_at |
AT1G64670
|
BDG1, BODYGUARD1, CED1, 9-CIS EPOXYCAROTENOID DIOXYGENASE DEFECTIVE 1 |
-0.514 |
0.003 |
| 326 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.513 |
0.040 |
| 327 |
262783_at |
AT1G10850
|
[Leucine-rich repeat protein kinase family protein] |
-0.509 |
-0.009 |
| 328 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.502 |
-0.071 |
| 329 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.502 |
0.040 |
| 330 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.500 |
0.008 |
| 331 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.491 |
0.031 |
| 332 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.490 |
0.057 |
| 333 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
-0.490 |
-0.033 |
| 334 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.486 |
0.019 |
| 335 |
245015_at |
ATCG00490
|
RBCL |
-0.485 |
-0.005 |
| 336 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.484 |
0.036 |
| 337 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
-0.483 |
0.041 |
| 338 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
-0.481 |
-0.005 |
| 339 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.480 |
0.036 |
| 340 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.477 |
-0.032 |
| 341 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.472 |
0.001 |
| 342 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.471 |
0.029 |
| 343 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.469 |
0.005 |
| 344 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.466 |
-0.030 |
| 345 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.466 |
0.053 |
| 346 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.464 |
0.012 |
| 347 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.464 |
-0.010 |
| 348 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-0.463 |
0.035 |
| 349 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.463 |
0.035 |
| 350 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.462 |
-0.001 |
| 351 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.458 |
0.047 |
| 352 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.455 |
0.160 |
| 353 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.454 |
-0.003 |
| 354 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.453 |
0.038 |
| 355 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.452 |
0.023 |
| 356 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.448 |
0.026 |
| 357 |
248105_at |
AT5G55280
|
ATFTSZ1-1, ARABIDOPSIS THALIANA HOMOLOG OF BACTERIAL CYTOKINESIS Z-RING PROTEIN FTSZ 1-1, CPFTSZ, CHLOROPLAST FTSZ, FTSZ1-1, homolog of bacterial cytokinesis Z-ring protein FTSZ 1-1 |
-0.448 |
0.015 |
| 358 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.447 |
-0.025 |
| 359 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.446 |
0.011 |
| 360 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.445 |
-0.042 |
| 361 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.444 |
0.047 |
| 362 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.444 |
0.044 |
| 363 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.441 |
0.009 |
| 364 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.441 |
-0.014 |
| 365 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.435 |
0.002 |
| 366 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.434 |
0.031 |
| 367 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.433 |
-0.010 |
| 368 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.431 |
-0.017 |
| 369 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.431 |
-0.010 |
| 370 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.422 |
0.099 |
| 371 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.420 |
0.026 |
| 372 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.419 |
0.049 |
| 373 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.419 |
0.016 |
| 374 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.418 |
-0.004 |
| 375 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
-0.416 |
0.036 |
| 376 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.415 |
0.020 |
| 377 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.414 |
0.069 |
| 378 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.414 |
-0.111 |
| 379 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.412 |
0.013 |
| 380 |
263829_at |
AT2G40435
|
unknown |
-0.411 |
-0.049 |
| 381 |
254181_at |
AT4G23940
|
ARC1, ACCUMULATION AND REPLICATION OF CHLOROPLASTS 1, FtsHi1, FTSH inactive protease 1 |
-0.406 |
-0.032 |
| 382 |
261363_at |
AT1G41830
|
SKS6, SKU5 SIMILAR 6, SKS6, SKU5-similar 6 |
-0.403 |
-0.005 |
| 383 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.402 |
0.011 |
| 384 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.400 |
0.085 |
| 385 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.399 |
-0.042 |
| 386 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.399 |
0.001 |
| 387 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.397 |
-0.009 |
| 388 |
264343_at |
AT1G11850
|
unknown |
-0.396 |
-0.048 |
| 389 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.396 |
0.003 |
| 390 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.396 |
0.004 |
| 391 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.394 |
0.032 |
| 392 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.393 |
-0.030 |
| 393 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.393 |
0.021 |
| 394 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
-0.392 |
0.033 |
| 395 |
261921_at |
AT1G65900
|
unknown |
-0.390 |
-0.052 |
| 396 |
247878_at |
AT5G57760
|
unknown |
-0.390 |
0.048 |
| 397 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.390 |
0.035 |
| 398 |
266244_at |
AT2G27740
|
unknown |
-0.389 |
0.054 |
| 399 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.389 |
-0.039 |
| 400 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.389 |
-0.009 |
| 401 |
253090_at |
AT4G36360
|
BGAL3, beta-galactosidase 3 |
-0.388 |
0.014 |
| 402 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.387 |
-0.010 |
| 403 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
-0.384 |
0.106 |
| 404 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
-0.384 |
0.014 |
| 405 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.383 |
0.028 |
| 406 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.383 |
-0.025 |
| 407 |
263488_at |
AT2G31840
|
MRL7-L, Mesophyll-cell RNAi Library line 7-like |
-0.382 |
-0.036 |
| 408 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.381 |
0.087 |
| 409 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.380 |
0.018 |
| 410 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.380 |
0.054 |
| 411 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.380 |
-0.001 |
| 412 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
-0.380 |
0.050 |
| 413 |
251319_at |
AT3G61610
|
[Galactose mutarotase-like superfamily protein] |
-0.376 |
-0.011 |
| 414 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
-0.375 |
0.050 |
| 415 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
-0.375 |
-0.003 |
| 416 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
-0.375 |
-0.010 |
| 417 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.375 |
0.009 |
| 418 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.375 |
0.021 |
| 419 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.374 |
-0.210 |
| 420 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.372 |
0.031 |
| 421 |
247068_at |
AT5G66800
|
unknown |
-0.370 |
0.006 |
| 422 |
248646_at |
AT5G49100
|
unknown |
-0.370 |
0.048 |
| 423 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
-0.369 |
-0.002 |
| 424 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.366 |
0.087 |
| 425 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.365 |
0.023 |
| 426 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.363 |
-0.054 |
| 427 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.362 |
0.031 |
| 428 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.362 |
-0.019 |
| 429 |
250337_at |
AT5G11790
|
NDL2, N-MYC downregulated-like 2 |
-0.361 |
0.050 |
| 430 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.361 |
0.008 |
| 431 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.361 |
0.016 |
| 432 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.360 |
-0.018 |
| 433 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.359 |
0.081 |
| 434 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
-0.359 |
-0.089 |
| 435 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.359 |
0.038 |
| 436 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.358 |
-0.007 |
| 437 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
-0.356 |
-0.016 |
| 438 |
250981_at |
AT5G03140
|
LecRK-VIII.2, L-type lectin receptor kinase VIII.2 |
-0.356 |
0.020 |
| 439 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.356 |
0.054 |
| 440 |
258480_at |
AT3G02640
|
unknown |
-0.355 |
0.025 |
| 441 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.354 |
0.003 |
| 442 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.354 |
0.045 |
| 443 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.353 |
0.030 |
| 444 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.352 |
0.015 |
| 445 |
259678_at |
AT1G77750
|
[Ribosomal protein S13/S18 family] |
-0.352 |
-0.001 |
| 446 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.351 |
-0.031 |
| 447 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
-0.351 |
-0.016 |
| 448 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.351 |
0.018 |
| 449 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.350 |
0.028 |
| 450 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.349 |
-0.040 |
| 451 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.349 |
-0.053 |
| 452 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.347 |
-0.011 |
| 453 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.347 |
0.002 |
| 454 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.347 |
-0.004 |
| 455 |
248623_at |
AT5G49170
|
unknown |
-0.347 |
0.031 |
| 456 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.346 |
-0.055 |
| 457 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
-0.344 |
0.003 |
| 458 |
247425_at |
AT5G62550
|
unknown |
-0.344 |
0.096 |
| 459 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.344 |
-0.042 |
| 460 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.343 |
0.006 |
| 461 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.343 |
0.021 |
| 462 |
247085_at |
AT5G66310
|
[ATP binding microtubule motor family protein] |
-0.341 |
0.047 |
| 463 |
256962_at |
AT3G13560
|
[O-Glycosyl hydrolases family 17 protein] |
-0.341 |
-0.005 |
| 464 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.341 |
-0.000 |
| 465 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.340 |
0.015 |
| 466 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.338 |
-0.015 |
| 467 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
-0.337 |
0.018 |
| 468 |
255642_at |
AT4G00820
|
iqd17, IQ-domain 17 |
-0.337 |
0.045 |
| 469 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.337 |
-0.001 |
| 470 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
-0.337 |
0.066 |
| 471 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
-0.336 |
0.064 |
| 472 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.335 |
0.033 |
| 473 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.335 |
0.145 |
| 474 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.335 |
-0.005 |
| 475 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.334 |
0.002 |
| 476 |
260857_at |
AT1G21880
|
LYM1, lysm domain GPI-anchored protein 1 precursor, LYP2, LysM-containing receptor protein 2 |
-0.334 |
0.017 |
| 477 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.333 |
-0.004 |
| 478 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.333 |
0.011 |
| 479 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.333 |
0.011 |
| 480 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.332 |
0.018 |
| 481 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.331 |
0.024 |
| 482 |
257608_at |
AT3G13860
|
HSP60-3A, heat shock protein 60-3A |
-0.331 |
0.001 |
| 483 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.331 |
0.034 |
| 484 |
261684_at |
AT1G47400
|
unknown |
-0.331 |
0.023 |
| 485 |
253607_at |
AT4G30330
|
[Small nuclear ribonucleoprotein family protein] |
-0.330 |
0.001 |
| 486 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.330 |
0.009 |
| 487 |
246511_at |
AT5G15490
|
UGD3, UDP-glucose dehydrogenase 3 |
-0.330 |
0.028 |
| 488 |
246425_at |
AT5G17420
|
ATCESA7, CESA7, CELLULOSE SYNTHASE CATALYTIC SUBUNIT 7, IRX3, IRREGULAR XYLEM 3, MUR10, MURUS 10 |
-0.330 |
0.038 |
| 489 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.330 |
0.032 |
| 490 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.330 |
-0.003 |
| 491 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.330 |
-0.009 |
| 492 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.329 |
0.002 |
| 493 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.329 |
0.043 |
| 494 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.329 |
-0.010 |
| 495 |
260667_at |
AT1G19440
|
KCS4, 3-ketoacyl-CoA synthase 4 |
-0.329 |
0.042 |
| 496 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.329 |
-0.002 |
| 497 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.329 |
-0.067 |
| 498 |
266079_at |
AT2G37860
|
LCD1, LOWER CELL DENSITY 1 |
-0.328 |
0.014 |
| 499 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
-0.328 |
-0.012 |
| 500 |
247881_at |
AT5G57700
|
[BNR/Asp-box repeat family protein] |
-0.328 |
0.003 |
| 501 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
-0.327 |
0.018 |
| 502 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.327 |
0.026 |
| 503 |
248139_at |
AT5G54970
|
unknown |
-0.326 |
-0.021 |
| 504 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.326 |
-0.007 |
| 505 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
-0.326 |
-0.084 |
| 506 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
-0.325 |
0.049 |
| 507 |
255460_at |
AT4G02800
|
unknown |
-0.323 |
0.063 |
| 508 |
266264_at |
AT2G27775
|
unknown |
-0.323 |
-0.053 |
| 509 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.323 |
0.001 |
| 510 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.321 |
0.033 |
| 511 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.321 |
0.042 |
| 512 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.320 |
0.019 |
| 513 |
247087_at |
AT5G66330
|
[Leucine-rich repeat (LRR) family protein] |
-0.320 |
0.029 |
| 514 |
247214_at |
AT5G64850
|
unknown |
-0.319 |
-0.015 |
| 515 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.319 |
0.053 |
| 516 |
264969_at |
AT1G67320
|
EMB2813, EMBRYO DEFECTIVE 2813 |
-0.319 |
-0.008 |
| 517 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.319 |
0.029 |
| 518 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.317 |
-0.005 |
| 519 |
247415_at |
AT5G63060
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.317 |
-0.015 |
| 520 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.316 |
0.004 |
| 521 |
248784_at |
AT5G47380
|
unknown |
-0.316 |
-0.009 |
| 522 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.316 |
0.026 |
| 523 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.316 |
0.042 |
| 524 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.316 |
-0.019 |
| 525 |
249167_at |
AT5G42860
|
unknown |
-0.315 |
0.056 |
| 526 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.315 |
0.014 |
| 527 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.315 |
-0.012 |
| 528 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.315 |
0.066 |
| 529 |
246772_at |
AT5G27490
|
[Integral membrane Yip1 family protein] |
-0.314 |
-0.028 |
| 530 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
-0.313 |
-0.007 |
| 531 |
251142_at |
AT5G01015
|
unknown |
-0.313 |
-0.027 |
| 532 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.312 |
0.053 |
| 533 |
254635_at |
AT4G18670
|
[Leucine-rich repeat (LRR) family protein] |
-0.312 |
0.056 |
| 534 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.312 |
0.034 |
| 535 |
262201_at |
AT2G01120
|
ATORC4, ORIGIN RECOGNITION COMPLEX SUBUNIT 4, ORC4, origin recognition complex subunit 4 |
-0.311 |
0.067 |
| 536 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.311 |
-0.001 |
| 537 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.311 |
0.192 |
| 538 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.311 |
-0.036 |
| 539 |
260009_at |
AT1G67950
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.310 |
-0.030 |
| 540 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
-0.310 |
-0.001 |
| 541 |
251249_at |
AT3G62160
|
[HXXXD-type acyl-transferase family protein] |
-0.310 |
0.003 |
| 542 |
256603_at |
AT3G28270
|
unknown |
-0.309 |
0.070 |
| 543 |
246415_at |
AT5G17160
|
unknown |
-0.309 |
0.056 |
| 544 |
267465_at |
AT2G19170
|
SLP3, subtilisin-like serine protease 3 |
-0.306 |
-0.015 |
| 545 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
-0.306 |
-0.014 |
| 546 |
245004_at |
ATCG00300
|
YCF9 |
-0.305 |
-0.010 |
| 547 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.305 |
0.006 |
| 548 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.305 |
0.004 |
| 549 |
251907_at |
AT3G53760
|
ATGCP4, GCP4, GAMMA-TUBULIN COMPLEX PROTEIN 4 |
-0.305 |
0.068 |
| 550 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
-0.305 |
-0.030 |
| 551 |
259710_at |
AT1G77670
|
[Pyridoxal phosphate (PLP)-dependent transferases superfamily protein] |
-0.303 |
0.022 |
| 552 |
253233_at |
AT4G34290
|
[SWIB/MDM2 domain superfamily protein] |
-0.303 |
-0.023 |
| 553 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
-0.303 |
-0.026 |
| 554 |
265464_at |
AT2G37080
|
RIP2, ROP interactive partner 2 |
-0.303 |
0.045 |
| 555 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.302 |
-0.010 |
| 556 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.302 |
0.024 |
| 557 |
263882_at |
AT2G21790
|
ATRNR1, RIBONUCLEOTIDE REDUCTASE LARGE SUBUNIT 1, CLS8, CRINKLY LEAVES 8, DPD2, defective in pollen organelle DNA degradation 2, R1, RIBONUCLEOTIDE REDUCTASE 1, RNR1, ribonucleotide reductase 1 |
-0.301 |
0.010 |
| 558 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.300 |
-0.026 |
| 559 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
-0.300 |
0.056 |
| 560 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.300 |
0.033 |
| 561 |
251183_at |
AT3G62630
|
unknown |
-0.300 |
0.012 |
| 562 |
259806_at |
AT1G47900
|
unknown |
-0.299 |
0.019 |
| 563 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.299 |
0.009 |
| 564 |
256043_at |
AT1G07210
|
[Ribosomal protein S18] |
-0.299 |
-0.016 |
| 565 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
-0.298 |
0.006 |
| 566 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.298 |
0.037 |
| 567 |
252089_at |
AT3G52110
|
unknown |
-0.298 |
0.002 |
| 568 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.298 |
-0.050 |
| 569 |
258549_at |
AT3G06930
|
ATPRMT4B, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 4B, PRMT4B, protein arginine methyltransferase 4B |
-0.298 |
-0.004 |
| 570 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.297 |
0.039 |
| 571 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.297 |
-0.023 |
| 572 |
262508_at |
AT1G11300
|
[protein serine/threonine kinases] |
-0.297 |
-0.054 |
| 573 |
262313_at |
AT1G70900
|
unknown |
-0.296 |
0.061 |
| 574 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
-0.296 |
0.027 |
| 575 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
-0.296 |
-0.050 |
| 576 |
254355_at |
AT4G22380
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
-0.296 |
-0.041 |
| 577 |
261439_at |
AT1G28395
|
unknown |
-0.295 |
0.011 |
| 578 |
261080_at |
AT1G07370
|
ATPCNA1, PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1, PCNA1, proliferating cellular nuclear antigen 1 |
-0.294 |
-0.006 |
| 579 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.294 |
0.041 |
| 580 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.294 |
0.053 |
| 581 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
-0.293 |
0.092 |
| 582 |
261292_at |
AT1G36940
|
unknown |
-0.292 |
0.094 |
| 583 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.292 |
0.001 |
| 584 |
254789_at |
AT4G12880
|
AtENODL19, ENODL19, early nodulin-like protein 19 |
-0.292 |
0.023 |
| 585 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
-0.292 |
0.081 |
| 586 |
263782_at |
AT2G46380
|
unknown |
-0.291 |
0.025 |
| 587 |
267037_at |
AT2G38320
|
TBL34, TRICHOME BIREFRINGENCE-LIKE 34 |
-0.291 |
0.116 |
| 588 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.289 |
0.059 |
| 589 |
263002_at |
AT1G54200
|
unknown |
-0.289 |
0.044 |
| 590 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-0.289 |
0.036 |
| 591 |
257533_at |
AT3G10840
|
[alpha/beta-Hydrolases superfamily protein] |
-0.288 |
-0.009 |
| 592 |
267586_at |
AT2G41950
|
unknown |
-0.288 |
-0.038 |
| 593 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.288 |
0.027 |
| 594 |
267220_at |
AT2G02500
|
ATMEPCT, ISPD, MCT, 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLTRANSFERASE |
-0.287 |
-0.016 |
| 595 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.286 |
0.041 |
| 596 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.285 |
0.036 |
| 597 |
246445_at |
AT5G17630
|
[Nucleotide/sugar transporter family protein] |
-0.285 |
0.016 |
| 598 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.285 |
0.024 |
| 599 |
245075_at |
AT2G23180
|
CYP96A1, cytochrome P450, family 96, subfamily A, polypeptide 1 |
-0.284 |
0.001 |
| 600 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
-0.284 |
0.028 |