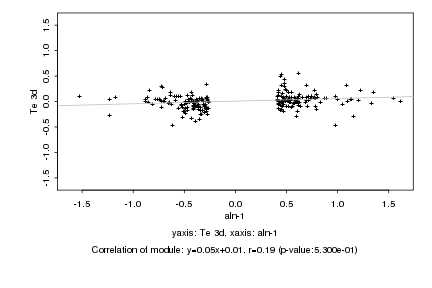
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
aln-1 |
Log2 signal ratio
Te 3d |
| 1 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
1.611 |
0.010 |
| 2 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
1.541 |
0.060 |
| 3 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
1.341 |
0.184 |
| 4 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.323 |
-0.028 |
| 5 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
1.214 |
0.233 |
| 6 |
263023_at |
AT1G23960
|
unknown |
1.203 |
0.038 |
| 7 |
265831_at |
AT2G14460
|
unknown |
1.152 |
-0.282 |
| 8 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
1.131 |
0.050 |
| 9 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
1.121 |
0.051 |
| 10 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
1.091 |
0.004 |
| 11 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
1.080 |
0.333 |
| 12 |
263673_at |
AT2G04800
|
unknown |
1.046 |
-0.041 |
| 13 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
0.991 |
0.053 |
| 14 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.974 |
-0.454 |
| 15 |
262010_at |
AT1G35612
|
[expressed protein] |
0.970 |
0.105 |
| 16 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
0.888 |
0.072 |
| 17 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
0.865 |
0.062 |
| 18 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.827 |
-0.013 |
| 19 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.796 |
0.086 |
| 20 |
249216_at |
AT5G42240
|
scpl42, serine carboxypeptidase-like 42 |
0.793 |
-0.138 |
| 21 |
256384_at |
AT1G66660
|
[Protein with RING/U-box and TRAF-like domains] |
0.790 |
0.143 |
| 22 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.783 |
-0.084 |
| 23 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.782 |
0.068 |
| 24 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
0.772 |
0.235 |
| 25 |
252432_at |
AT3G47680
|
[DNA binding] |
0.771 |
0.066 |
| 26 |
245444_at |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
0.767 |
0.087 |
| 27 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.741 |
0.109 |
| 28 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
0.736 |
0.118 |
| 29 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.734 |
0.069 |
| 30 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
0.706 |
0.104 |
| 31 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.706 |
0.034 |
| 32 |
248079_at |
AT5G55790
|
unknown |
0.699 |
-0.089 |
| 33 |
253074_at |
AT4G36140
|
[disease resistance protein (TIR-NBS-LRR class), putative] |
0.695 |
0.038 |
| 34 |
246888_at |
AT5G26270
|
unknown |
0.688 |
0.086 |
| 35 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.687 |
0.318 |
| 36 |
254852_at |
AT4G12020
|
ATWRKY19, MAPKKK11, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 11, MEKK4, MAPK/ERK KINASE KINASE 4, WRKY19 |
0.677 |
-0.018 |
| 37 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
0.654 |
0.095 |
| 38 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
0.629 |
-0.081 |
| 39 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
0.626 |
-0.001 |
| 40 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.622 |
0.142 |
| 41 |
253827_at |
AT4G28085
|
unknown |
0.616 |
0.555 |
| 42 |
245079_at |
AT2G23330
|
[copia-like retrotransposon family, has a 3.9e-195 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
0.615 |
-0.002 |
| 43 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.614 |
0.074 |
| 44 |
264824_at |
AT1G03420
|
Sadhu4-2, sadhu non-coding retrotransposon 4-2 |
0.612 |
0.099 |
| 45 |
252937_at |
AT4G39180
|
ATSEC14, ARABIDOPSIS THALIANA SECRETION 14, SEC14, SECRETION 14 |
0.610 |
-0.075 |
| 46 |
256114_at |
AT1G16850
|
unknown |
0.605 |
0.069 |
| 47 |
248936_at |
AT5G45710
|
AT-HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, RHA1, ROOT HANDEDNESS 1 |
0.605 |
-0.019 |
| 48 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
0.605 |
-0.007 |
| 49 |
249904_at |
AT5G22700
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.603 |
-0.184 |
| 50 |
255527_at |
AT4G02360
|
unknown |
0.597 |
-0.290 |
| 51 |
263083_at |
AT2G27190
|
ATPAP1, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 1, ATPAP12, PURPLE ACID PHOSPHATASE 12, PAP1, PURPLE ACID PHOSPHATASE 1, PAP12, purple acid phosphatase 12 |
0.591 |
-0.005 |
| 52 |
245169_at |
AT2G33220
|
[GRIM-19 protein] |
0.588 |
0.011 |
| 53 |
263883_at |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.584 |
0.039 |
| 54 |
262362_at |
AT1G72840
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.581 |
0.068 |
| 55 |
256211_at |
AT1G50960
|
ATGA2OX7, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 7, GA2OX7, gibberellin 2-oxidase 7 |
0.571 |
0.082 |
| 56 |
255751_at |
AT1G31950
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.569 |
-0.020 |
| 57 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
0.567 |
-0.025 |
| 58 |
264454_at |
AT1G10320
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.554 |
-0.087 |
| 59 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
0.547 |
0.045 |
| 60 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.544 |
0.187 |
| 61 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.540 |
-0.104 |
| 62 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
0.539 |
0.087 |
| 63 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
0.535 |
-0.001 |
| 64 |
259755_at |
AT1G71070
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.530 |
0.030 |
| 65 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.527 |
-0.012 |
| 66 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
0.524 |
-0.017 |
| 67 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
0.521 |
0.051 |
| 68 |
248954_at |
AT5G45420
|
maMYB, membrane anchored MYB |
0.514 |
0.049 |
| 69 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.514 |
0.196 |
| 70 |
256764_at |
AT3G29310
|
[calmodulin-binding protein-related] |
0.512 |
-0.087 |
| 71 |
259653_at |
AT1G55240
|
unknown |
0.512 |
0.037 |
| 72 |
256493_at |
AT1G31600
|
AtTRM9, Arabidopsis thaliana tRNA methyltransferase 9, TRM9, tRNA methyltransferase 9 |
0.504 |
0.024 |
| 73 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.503 |
0.022 |
| 74 |
260557_at |
AT2G43610
|
[Chitinase family protein] |
0.494 |
0.117 |
| 75 |
258544_at |
AT3G07040
|
RPM1, RESISTANCE TO P. SYRINGAE PV MACULICOLA 1, RPS3, RESISTANCE TO PSEUDOMONAS SYRINGAE 3 |
0.493 |
0.218 |
| 76 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
0.490 |
-0.080 |
| 77 |
257554_at |
AT3G24890
|
ATVAMP728, VAMP728, vesicle-associated membrane protein 728 |
0.481 |
0.009 |
| 78 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.481 |
0.028 |
| 79 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.481 |
0.246 |
| 80 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.474 |
0.067 |
| 81 |
246235_at |
AT4G36830
|
HOS3-1 |
0.473 |
0.313 |
| 82 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.473 |
0.370 |
| 83 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.473 |
0.440 |
| 84 |
255702_at |
AT4G00230
|
XSP1, xylem serine peptidase 1 |
0.470 |
0.091 |
| 85 |
248977_at |
AT5G45020
|
[Glutathione S-transferase family protein] |
0.468 |
0.010 |
| 86 |
257497_at |
AT1G51430
|
unknown |
0.464 |
-0.180 |
| 87 |
265907_at |
AT2G25650
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
0.463 |
-0.048 |
| 88 |
252444_at |
AT3G46980
|
PHT4;3, phosphate transporter 4;3 |
0.462 |
0.040 |
| 89 |
262496_at |
AT1G21790
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
0.462 |
0.020 |
| 90 |
246394_at |
AT1G58160
|
JAX1, JACALIN-TYPE LECTIN REQUIRED FOR POTEXVIRUS RESISTANCE 1 |
0.454 |
0.027 |
| 91 |
254586_at |
AT4G19510
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.453 |
-0.004 |
| 92 |
254965_at |
AT4G11090
|
TBL23, TRICHOME BIREFRINGENCE-LIKE 23 |
0.452 |
0.035 |
| 93 |
246905_at |
AT5G25570
|
unknown |
0.451 |
-0.080 |
| 94 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.451 |
0.162 |
| 95 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.449 |
-0.014 |
| 96 |
251207_at |
AT3G63050
|
unknown |
0.449 |
0.541 |
| 97 |
266108_at |
AT2G37900
|
[Major facilitator superfamily protein] |
0.448 |
-0.153 |
| 98 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
0.447 |
0.111 |
| 99 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
0.443 |
0.316 |
| 100 |
267185_at |
AT2G43950
|
ATOEP37, ARABIDOPSIS CHLOROPLAST OUTER ENVELOPE PROTEIN 37, OEP37, chloroplast outer envelope protein 37 |
0.440 |
-0.063 |
| 101 |
259796_at |
AT1G64270
|
[Mutator-like transposase family, has a 1.0e-06 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.437 |
-0.166 |
| 102 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.433 |
0.499 |
| 103 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.422 |
0.003 |
| 104 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.419 |
0.126 |
| 105 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
0.419 |
-0.119 |
| 106 |
255005_at |
AT4G09990
|
GXM2, glucuronoxylan methyltransferase 2 |
0.418 |
-0.026 |
| 107 |
245453_at |
AT4G16900
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.416 |
0.012 |
| 108 |
249619_at |
AT5G37500
|
GORK, gated outwardly-rectifying K+ channel |
0.414 |
0.194 |
| 109 |
256568_at |
AT3G19520
|
unknown |
0.413 |
-0.041 |
| 110 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.412 |
0.228 |
| 111 |
249185_at |
AT5G43030
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.409 |
0.022 |
| 112 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
0.409 |
0.117 |
| 113 |
257353_at |
AT2G23050
|
MEL4, MAB4/ENP/NPY1-LIKE 4, NPY4, NAKED PINS IN YUC MUTANTS 4 |
0.409 |
0.054 |
| 114 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
-1.531 |
0.101 |
| 115 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-1.241 |
0.056 |
| 116 |
252345_at |
AT3G48640
|
unknown |
-1.234 |
-0.257 |
| 117 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
-1.181 |
0.097 |
| 118 |
250135_at |
AT5G15360
|
unknown |
-0.883 |
0.055 |
| 119 |
258028_at |
AT3G27473
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.881 |
0.003 |
| 120 |
254574_at |
AT4G19430
|
unknown |
-0.869 |
0.081 |
| 121 |
262421_at |
AT1G50290
|
unknown |
-0.852 |
-0.002 |
| 122 |
256631_at |
AT3G28320
|
unknown |
-0.848 |
0.233 |
| 123 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.813 |
-0.055 |
| 124 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.789 |
0.046 |
| 125 |
246857_at |
AT5G25920
|
unknown |
-0.765 |
0.052 |
| 126 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.749 |
0.049 |
| 127 |
252347_at |
AT3G48130
|
RSU1 |
-0.739 |
0.036 |
| 128 |
244999_at |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
-0.730 |
0.013 |
| 129 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.729 |
0.300 |
| 130 |
265893_at |
AT2G15042
|
[Leucine-rich repeat (LRR) family protein] |
-0.724 |
-0.109 |
| 131 |
258016_at |
AT3G19350
|
MPC, maternally expressed pab C-terminal |
-0.720 |
0.282 |
| 132 |
256382_at |
AT1G66860
|
[Class I glutamine amidotransferase-like superfamily protein] |
-0.704 |
0.018 |
| 133 |
262651_at |
AT1G14100
|
FUT8, fucosyltransferase 8 |
-0.696 |
0.006 |
| 134 |
246168_at |
AT5G32460
|
[Transcriptional factor B3 family protein] |
-0.685 |
0.068 |
| 135 |
265063_at |
AT1G61500
|
[S-locus lectin protein kinase family protein] |
-0.663 |
-0.031 |
| 136 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
-0.650 |
-0.019 |
| 137 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-0.641 |
0.185 |
| 138 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.636 |
0.123 |
| 139 |
256859_at |
AT3G22940
|
[F-box associated ubiquitination effector family protein] |
-0.635 |
-0.044 |
| 140 |
255879_at |
AT1G67000
|
[Protein kinase superfamily protein] |
-0.622 |
-0.469 |
| 141 |
257479_at |
AT1G16150
|
WAKL4, wall associated kinase-like 4 |
-0.604 |
0.118 |
| 142 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.594 |
0.034 |
| 143 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.579 |
0.114 |
| 144 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
-0.565 |
-0.121 |
| 145 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-0.559 |
0.102 |
| 146 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.545 |
0.113 |
| 147 |
264167_at |
AT1G02060
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.534 |
-0.075 |
| 148 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.533 |
-0.044 |
| 149 |
252717_at |
AT3G43930
|
[BRCT domain-containing DNA repair protein] |
-0.524 |
-0.101 |
| 150 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.518 |
-0.310 |
| 151 |
255653_at |
AT4G00960
|
[Protein kinase superfamily protein] |
-0.515 |
-0.189 |
| 152 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.505 |
-0.054 |
| 153 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
-0.502 |
-0.196 |
| 154 |
261302_at |
AT1G48580
|
unknown |
-0.498 |
-0.228 |
| 155 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
-0.492 |
0.007 |
| 156 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.489 |
-0.123 |
| 157 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.487 |
-0.033 |
| 158 |
253689_at |
AT4G29770
|
unknown |
-0.477 |
-0.158 |
| 159 |
262719_at |
AT1G43590
|
unknown |
-0.476 |
0.128 |
| 160 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.471 |
-0.101 |
| 161 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.468 |
0.041 |
| 162 |
253532_at |
AT4G31570
|
unknown |
-0.466 |
0.018 |
| 163 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.459 |
-0.010 |
| 164 |
266827_at |
AT2G22920
|
SCPL12, serine carboxypeptidase-like 12 |
-0.458 |
-0.012 |
| 165 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.446 |
0.066 |
| 166 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
-0.439 |
-0.331 |
| 167 |
262665_at |
AT1G14070
|
FUT7, fucosyltransferase 7 |
-0.433 |
0.054 |
| 168 |
266412_at |
AT2G38720
|
MAP65-5, microtubule-associated protein 65-5 |
-0.432 |
0.005 |
| 169 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.431 |
0.189 |
| 170 |
258147_at |
AT3G18070
|
BGLU43, beta glucosidase 43 |
-0.427 |
-0.000 |
| 171 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
-0.422 |
0.130 |
| 172 |
253945_at |
AT4G27050
|
[F-box/RNI-like superfamily protein] |
-0.418 |
-0.064 |
| 173 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.417 |
-0.126 |
| 174 |
257139_at |
AT3G28890
|
AtRLP43, receptor like protein 43, RLP43, receptor like protein 43 |
-0.416 |
-0.080 |
| 175 |
262529_at |
AT1G17250
|
AtRLP3, receptor like protein 3, RFO2, RESISTANCE TO FUSARIUM OXYSPORUM 2, RLP3, receptor like protein 3 |
-0.412 |
0.016 |
| 176 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.411 |
-0.154 |
| 177 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
-0.404 |
-0.003 |
| 178 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
-0.404 |
-0.038 |
| 179 |
260799_at |
AT1G78270
|
AtUGT85A4, UDP-glucosyl transferase 85A4, UGT85A4, UDP-glucosyl transferase 85A4 |
-0.401 |
-0.099 |
| 180 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
-0.398 |
-0.009 |
| 181 |
256603_at |
AT3G28270
|
unknown |
-0.391 |
-0.376 |
| 182 |
248015_at |
AT5G56370
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.391 |
-0.074 |
| 183 |
258013_at |
AT3G19320
|
[Leucine-rich repeat (LRR) family protein] |
-0.388 |
0.041 |
| 184 |
255627_at |
AT4G00955
|
unknown |
-0.376 |
-0.067 |
| 185 |
248991_at |
AT5G45220
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.375 |
0.015 |
| 186 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
-0.375 |
0.006 |
| 187 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.375 |
0.039 |
| 188 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.373 |
-0.086 |
| 189 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.371 |
-0.099 |
| 190 |
260464_at |
AT1G10920
|
LOV1, LOCUS ORCHESTRATING VICTORIN EFFECTS1 |
-0.369 |
-0.045 |
| 191 |
262126_at |
AT1G59620
|
CW9 |
-0.368 |
-0.115 |
| 192 |
257764_at |
AT3G23010
|
AtRLP36, receptor like protein 36, RLP36, receptor like protein 36 |
-0.366 |
-0.144 |
| 193 |
250596_at |
AT5G07780
|
AtFH19, FH19, formin homolog 19 |
-0.363 |
-0.093 |
| 194 |
253940_at |
AT4G26950
|
unknown |
-0.360 |
-0.336 |
| 195 |
255317_at |
AT4G04180
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.353 |
0.072 |
| 196 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.349 |
-0.101 |
| 197 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
-0.349 |
-0.164 |
| 198 |
254145_at |
AT4G24700
|
unknown |
-0.347 |
-0.245 |
| 199 |
266164_at |
AT2G28050
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.339 |
0.024 |
| 200 |
265356_at |
AT2G16595
|
[Translocon-associated protein (TRAP), alpha subunit] |
-0.336 |
0.061 |
| 201 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.335 |
-0.239 |
| 202 |
252712_at |
AT3G43800
|
ATGSTU27, glutathione S-transferase tau 27, GSTU27, glutathione S-transferase tau 27 |
-0.329 |
0.045 |
| 203 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
-0.329 |
-0.163 |
| 204 |
257929_at |
AT3G16980
|
NRPB9A, NRPD9A, NRPE9A |
-0.320 |
-0.050 |
| 205 |
253211_at |
AT4G34880
|
[Amidase family protein] |
-0.317 |
0.005 |
| 206 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.315 |
0.015 |
| 207 |
260060_at |
AT1G73680
|
ALPHA DOX2, alpha dioxygenase, alpha-DOX2, alpha-dioxygenase 2 |
-0.312 |
-0.096 |
| 208 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
-0.310 |
-0.215 |
| 209 |
258198_at |
AT3G14020
|
NF-YA6, nuclear factor Y, subunit A6 |
-0.309 |
-0.059 |
| 210 |
252707_at |
AT3G43790
|
ZIFL2, zinc induced facilitator-like 2 |
-0.296 |
-0.056 |
| 211 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
-0.295 |
-0.125 |
| 212 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
-0.292 |
0.070 |
| 213 |
259753_at |
AT1G71050
|
HIPP20, heavy metal associated isoprenylated plant protein 20 |
-0.291 |
-0.138 |
| 214 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.290 |
0.097 |
| 215 |
258546_at |
AT3G07060
|
emb1974, embryo defective 1974 |
-0.289 |
-0.112 |
| 216 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.285 |
0.342 |
| 217 |
263546_at |
AT2G21550
|
[Bifunctional dihydrofolate reductase/thymidylate synthase] |
-0.284 |
-0.196 |
| 218 |
256767_at |
AT3G13680
|
[F-box and associated interaction domains-containing protein] |
-0.283 |
0.059 |
| 219 |
260211_at |
AT1G74440
|
unknown |
-0.282 |
-0.249 |
| 220 |
262074_at |
AT1G59670
|
ATGSTU15, glutathione S-transferase TAU 15, GSTU15, glutathione S-transferase TAU 15 |
-0.282 |
0.021 |
| 221 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.278 |
-0.127 |
| 222 |
246438_at |
AT5G17580
|
[Phototropic-responsive NPH3 family protein] |
-0.274 |
0.081 |
| 223 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.272 |
-0.017 |
| 224 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
-0.271 |
0.017 |
| 225 |
254548_at |
AT4G19865
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.270 |
-0.122 |