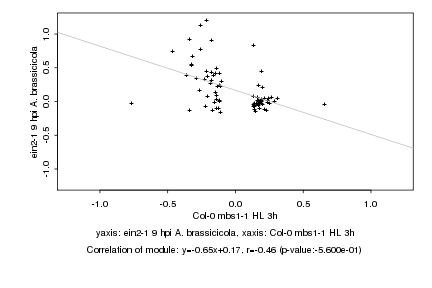
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 mbs1-1 HL 3h |
Log2 signal ratio
ein2-1 9 hpi A. brassicicola |
| 1 |
252036_at |
AT3G52070
|
unknown |
0.650 |
-0.032 |
| 2 |
258177_at |
AT3G21660
|
[UBX domain-containing protein] |
0.309 |
0.051 |
| 3 |
252224_at |
AT3G49860
|
ARLA1B, ADP-ribosylation factor-like A1B, ATARLA1B, ADP-ribosylation factor-like A1B |
0.288 |
0.002 |
| 4 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.265 |
0.071 |
| 5 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.248 |
-0.016 |
| 6 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.240 |
0.048 |
| 7 |
258960_at |
AT3G10590
|
[Duplicated homeodomain-like superfamily protein] |
0.234 |
0.029 |
| 8 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
0.230 |
-0.014 |
| 9 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
0.227 |
-0.125 |
| 10 |
261088_at |
AT1G07590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.209 |
-0.111 |
| 11 |
253155_at |
AT4G35720
|
unknown |
0.208 |
0.057 |
| 12 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.195 |
-0.039 |
| 13 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.195 |
0.218 |
| 14 |
259084_at |
AT3G04800
|
ATTIM23-3, translocase inner membrane subunit 23-3, TIM23-3, translocase inner membrane subunit 23-3 |
0.191 |
0.030 |
| 15 |
255530_at |
AT4G02140
|
unknown |
0.191 |
-0.027 |
| 16 |
261960_at |
AT1G36550
|
[similar to retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa (japonica cultivar-group)] (GB:ABA98367.2)] |
0.187 |
0.005 |
| 17 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.186 |
0.027 |
| 18 |
251689_at |
AT3G56500
|
[serine-rich protein-related] |
0.186 |
0.455 |
| 19 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
0.174 |
-0.101 |
| 20 |
253595_at |
AT4G30830
|
unknown |
0.174 |
-0.029 |
| 21 |
245495_at |
AT4G16400
|
unknown |
0.171 |
-0.006 |
| 22 |
251377_at |
AT3G60650
|
unknown |
0.170 |
-0.016 |
| 23 |
264468_at |
AT1G10310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.165 |
-0.029 |
| 24 |
262795_at |
AT1G13130
|
[Cellulase (glycosyl hydrolase family 5) protein] |
0.165 |
-0.045 |
| 25 |
249617_at |
AT5G37450
|
[Leucine-rich repeat protein kinase family protein] |
0.163 |
0.003 |
| 26 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.163 |
0.248 |
| 27 |
257094_at |
AT3G20480
|
tetraacyldisaccharide 4'-kinase family protein |
0.163 |
0.026 |
| 28 |
245126_at |
AT2G47460
|
ATMYB12, MYB DOMAIN PROTEIN 12, MYB12, myb domain protein 12, PFG1, PRODUCTION OF FLAVONOL GLYCOSIDES 1 |
0.162 |
0.020 |
| 29 |
258619_at |
AT3G02780
|
IDI2, ATISOPENTENYL DIPHOSPHE ISOMERASE 2, IPIAT1, IPP2, isopentenyl pyrophosphate:dimethylallyl pyrophosphate isomerase 2 |
0.160 |
-0.034 |
| 30 |
262904_at |
AT1G59725
|
[DNAJ heat shock family protein] |
0.156 |
0.072 |
| 31 |
245419_at |
AT4G17380
|
ATMSH4, ARABIDOPSIS MUTS HOMOLOG 4, MSH4, MUTS HOMOLOG 4, MSH4, MUTS-like protein 4 |
0.148 |
-0.027 |
| 32 |
255943_at |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
0.144 |
-0.137 |
| 33 |
252915_at |
AT4G38810
|
[Calcium-binding EF-hand family protein] |
0.139 |
-0.057 |
| 34 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
0.137 |
-0.064 |
| 35 |
254295_at |
AT4G23080
|
unknown |
0.137 |
-0.050 |
| 36 |
257505_at |
AT1G47940
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.135 |
-0.019 |
| 37 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.134 |
-0.114 |
| 38 |
262815_at |
AT1G11610
|
CYP71A18, cytochrome P450, family 71, subfamily A, polypeptide 18 |
0.130 |
0.834 |
| 39 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.128 |
-0.069 |
| 40 |
246592_at |
AT5G14890
|
[NHL domain-containing protein] |
0.127 |
0.082 |
| 41 |
267478_at |
AT2G02700
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.126 |
-0.044 |
| 42 |
258623_at |
AT3G02790
|
MBS1, METHYLENE BLUE SENSITIVITY 1 |
-0.773 |
-0.027 |
| 43 |
263182_at |
AT1G05575
|
unknown |
-0.470 |
0.746 |
| 44 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
-0.365 |
0.387 |
| 45 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.346 |
-0.128 |
| 46 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
-0.346 |
0.924 |
| 47 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.326 |
0.538 |
| 48 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.325 |
0.562 |
| 49 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
-0.318 |
0.672 |
| 50 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.294 |
0.347 |
| 51 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.266 |
0.167 |
| 52 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
-0.263 |
0.777 |
| 53 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.263 |
1.132 |
| 54 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.233 |
0.340 |
| 55 |
264738_at |
AT1G62250
|
unknown |
-0.224 |
-0.062 |
| 56 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.217 |
0.458 |
| 57 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.215 |
1.215 |
| 58 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
-0.211 |
0.075 |
| 59 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.211 |
0.382 |
| 60 |
258117_at |
AT3G14700
|
[SART-1 family] |
-0.186 |
0.277 |
| 61 |
258577_at |
AT3G04220
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.184 |
0.316 |
| 62 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
-0.181 |
0.431 |
| 63 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.180 |
0.907 |
| 64 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.171 |
-0.121 |
| 65 |
252851_at |
AT4G40080
|
[ENTH/ANTH/VHS superfamily protein] |
-0.165 |
0.390 |
| 66 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.159 |
-0.004 |
| 67 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.154 |
0.419 |
| 68 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.153 |
0.139 |
| 69 |
256509_at |
AT1G75300
|
[NmrA-like negative transcriptional regulator family protein] |
-0.147 |
-0.091 |
| 70 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
-0.144 |
0.493 |
| 71 |
253149_at |
AT4G35650
|
IDH-III, isocitrate dehydrogenase III |
-0.142 |
0.101 |
| 72 |
249905_at |
AT5G22720
|
[F-box/RNI-like superfamily protein] |
-0.141 |
0.034 |
| 73 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
-0.138 |
0.236 |
| 74 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.130 |
-0.101 |
| 75 |
255505_at |
AT4G02080
|
ASAR1, ATSAR2, secretion-associated RAS super family 2, ATSARA1C, SAR2, secretion-associated RAS super family 2 |
-0.125 |
0.019 |
| 76 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
-0.121 |
0.008 |
| 77 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
-0.120 |
0.246 |
| 78 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
-0.119 |
0.428 |
| 79 |
248976_at |
AT5G44930
|
ARAD2, ARABINAN DEFICIENT 2 |
-0.115 |
-0.162 |
| 80 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
-0.112 |
0.232 |
| 81 |
257074_at |
AT3G19660
|
unknown |
-0.110 |
0.307 |