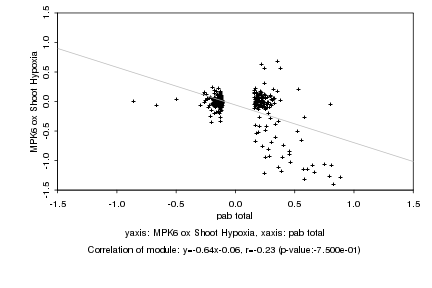
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pab total |
Log2 signal ratio
MPK6 ox Shoot Hypoxia |
| 1 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
0.882 |
-1.284 |
| 2 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
0.821 |
-1.389 |
| 3 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.803 |
-1.071 |
| 4 |
257057_at |
AT3G15310
|
unknown |
0.799 |
-0.046 |
| 5 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
0.789 |
-1.264 |
| 6 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
0.746 |
-1.058 |
| 7 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
0.666 |
-1.197 |
| 8 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
0.650 |
-1.080 |
| 9 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.601 |
-1.139 |
| 10 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.580 |
-0.257 |
| 11 |
266393_at |
AT2G41260
|
ATM17, M17 |
0.577 |
-1.311 |
| 12 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
0.569 |
-1.151 |
| 13 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.552 |
-0.660 |
| 14 |
256569_at |
AT3G19550
|
unknown |
0.528 |
0.210 |
| 15 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.519 |
-0.502 |
| 16 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
0.463 |
-1.030 |
| 17 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
0.453 |
-0.886 |
| 18 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.452 |
-0.844 |
| 19 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.399 |
-0.731 |
| 20 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.394 |
-0.938 |
| 21 |
250468_at |
AT5G10120
|
[Ethylene insensitive 3 family protein] |
0.385 |
-1.171 |
| 22 |
256096_at |
AT1G13650
|
unknown |
0.379 |
0.024 |
| 23 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.377 |
0.565 |
| 24 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.361 |
-1.103 |
| 25 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.359 |
-0.322 |
| 26 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.354 |
0.186 |
| 27 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.347 |
0.685 |
| 28 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.336 |
-0.593 |
| 29 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.332 |
-0.381 |
| 30 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.329 |
0.064 |
| 31 |
262719_at |
AT1G43590
|
unknown |
0.328 |
-0.032 |
| 32 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
0.319 |
0.053 |
| 33 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.318 |
0.043 |
| 34 |
249127_at |
AT5G43500
|
ARP9, actin-related protein 9, ATARP9, actin-related protein 9 |
0.314 |
0.210 |
| 35 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.310 |
0.028 |
| 36 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.304 |
0.040 |
| 37 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.302 |
-0.691 |
| 38 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.296 |
-0.066 |
| 39 |
257502_at |
AT1G78110
|
unknown |
0.292 |
-0.049 |
| 40 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.290 |
-0.278 |
| 41 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.290 |
0.090 |
| 42 |
263545_at |
AT2G21560
|
unknown |
0.288 |
0.110 |
| 43 |
260859_at |
AT1G43780
|
scpl44, serine carboxypeptidase-like 44 |
0.286 |
-0.096 |
| 44 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
0.281 |
-0.925 |
| 45 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.278 |
-0.200 |
| 46 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.274 |
-0.005 |
| 47 |
250624_at |
AT5G07330
|
unknown |
0.274 |
-0.807 |
| 48 |
251772_at |
AT3G55920
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.269 |
-0.012 |
| 49 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
0.266 |
-0.101 |
| 50 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.262 |
0.069 |
| 51 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.262 |
-0.046 |
| 52 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
0.257 |
-0.038 |
| 53 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
0.254 |
-0.422 |
| 54 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.253 |
-0.129 |
| 55 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.253 |
0.116 |
| 56 |
258935_at |
AT3G10120
|
unknown |
0.252 |
-0.114 |
| 57 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
0.251 |
-0.934 |
| 58 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.250 |
0.053 |
| 59 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.250 |
-0.488 |
| 60 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
0.245 |
-1.209 |
| 61 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
0.241 |
-0.042 |
| 62 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.240 |
-0.004 |
| 63 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.240 |
0.561 |
| 64 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
0.238 |
0.127 |
| 65 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.237 |
-0.068 |
| 66 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.237 |
0.316 |
| 67 |
254100_at |
AT4G25020
|
[D111/G-patch domain-containing protein] |
0.235 |
-0.025 |
| 68 |
256926_at |
AT3G22540
|
unknown |
0.235 |
0.085 |
| 69 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
0.233 |
-0.080 |
| 70 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
0.233 |
0.007 |
| 71 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.225 |
-0.761 |
| 72 |
251050_at |
AT5G02440
|
unknown |
0.224 |
-0.005 |
| 73 |
245725_at |
AT1G73370
|
ATSUS6, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 6, SUS6, sucrose synthase 6 |
0.223 |
0.020 |
| 74 |
254390_at |
AT4G21940
|
CPK15, calcium-dependent protein kinase 15 |
0.222 |
0.102 |
| 75 |
246817_at |
AT5G27240
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.221 |
-0.094 |
| 76 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.218 |
0.641 |
| 77 |
249136_at |
AT5G43180
|
unknown |
0.215 |
-0.053 |
| 78 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
0.213 |
0.156 |
| 79 |
262081_at |
AT1G59540
|
ZCF125 |
0.213 |
-0.017 |
| 80 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.213 |
0.073 |
| 81 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.212 |
0.068 |
| 82 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
0.212 |
0.007 |
| 83 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.212 |
0.103 |
| 84 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
0.210 |
-0.003 |
| 85 |
261031_at |
AT1G17360
|
unknown |
0.210 |
-0.016 |
| 86 |
253978_at |
AT4G26660
|
unknown |
0.209 |
-0.084 |
| 87 |
267166_at |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
0.206 |
0.181 |
| 88 |
261832_at |
AT1G10650
|
[SBP (S-ribonuclease binding protein) family protein] |
0.206 |
0.140 |
| 89 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.205 |
-0.037 |
| 90 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.202 |
-0.420 |
| 91 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.201 |
0.029 |
| 92 |
248814_at |
AT5G46910
|
[Transcription factor jumonji (jmj) family protein / zinc finger (C5HC2 type) family protein] |
0.200 |
0.128 |
| 93 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.197 |
-0.263 |
| 94 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.197 |
0.052 |
| 95 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.194 |
0.025 |
| 96 |
247340_at |
AT5G63700
|
[zinc ion binding] |
0.194 |
0.106 |
| 97 |
246415_at |
AT5G17160
|
unknown |
0.194 |
-0.060 |
| 98 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.194 |
-0.509 |
| 99 |
260367_at |
AT1G69760
|
unknown |
0.194 |
0.069 |
| 100 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
0.193 |
-0.092 |
| 101 |
248015_at |
AT5G56370
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.191 |
0.059 |
| 102 |
265993_at |
AT2G24160
|
[pseudogene] |
0.190 |
0.015 |
| 103 |
253243_at |
AT4G34560
|
unknown |
0.190 |
-0.131 |
| 104 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
0.182 |
0.019 |
| 105 |
263607_at |
AT2G16270
|
unknown |
0.181 |
-0.012 |
| 106 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.181 |
0.033 |
| 107 |
248889_at |
AT5G46230
|
unknown |
0.180 |
-0.104 |
| 108 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
0.179 |
0.105 |
| 109 |
248150_at |
AT5G54670
|
ATK3, kinesin 3, KATC, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA C |
0.178 |
0.094 |
| 110 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
0.176 |
-0.042 |
| 111 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
0.175 |
-0.527 |
| 112 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
0.174 |
-0.027 |
| 113 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
0.174 |
0.069 |
| 114 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
0.173 |
-0.067 |
| 115 |
248819_at |
AT5G47050
|
[SBP (S-ribonuclease binding protein) family protein] |
0.172 |
0.152 |
| 116 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
0.171 |
-0.058 |
| 117 |
252077_at |
AT3G51720
|
unknown |
0.171 |
0.026 |
| 118 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
0.171 |
0.008 |
| 119 |
264001_at |
AT2G22420
|
[Peroxidase superfamily protein] |
0.170 |
0.003 |
| 120 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.170 |
0.036 |
| 121 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
0.170 |
0.160 |
| 122 |
267037_at |
AT2G38320
|
TBL34, TRICHOME BIREFRINGENCE-LIKE 34 |
0.169 |
0.017 |
| 123 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
0.167 |
-0.677 |
| 124 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.166 |
-0.036 |
| 125 |
247990_at |
AT5G56360
|
PSL4, PRIORITY IN SWEET LIFE 4 |
0.166 |
-0.004 |
| 126 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.165 |
-0.030 |
| 127 |
256721_at |
AT2G34150
|
ATRANGAP2, ATSCAR1, SCAR1, WAVE1, WISKOTT-ALDRICH SYNDROME PROTEIN FAMILY VERPROLIN HOMOLOGOUS PROTEIN 1 |
0.164 |
0.044 |
| 128 |
261100_at |
AT1G63020
|
NRPD1, NRPD1A, nuclear RNA polymerase D1A, POL IVA, NUCLEAR RNA POLYMERASE D 1A, SDE4, SMD2, SILENCING MOVEMENT DEFICIENT 2 |
0.163 |
0.036 |
| 129 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.162 |
-0.068 |
| 130 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.162 |
0.225 |
| 131 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
0.162 |
-0.018 |
| 132 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.162 |
-0.393 |
| 133 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.157 |
-0.039 |
| 134 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.156 |
0.168 |
| 135 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
0.156 |
0.144 |
| 136 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
0.155 |
0.197 |
| 137 |
261471_at |
AT1G14460
|
[AAA-type ATPase family protein] |
0.154 |
-0.109 |
| 138 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.154 |
0.071 |
| 139 |
253280_at |
AT4G34110
|
ATPAB2, ARABIDOPSIS POLY(A) BINDING 2, PAB2, poly(A) binding protein 2, PABP2, POLY(A) BINDING PROTEIN 2 |
-0.864 |
0.008 |
| 140 |
261614_at |
AT1G49760
|
PAB8, poly(A) binding protein 8, PABP8, POLY(A) BINDING PROTEIN 8 |
-0.672 |
-0.062 |
| 141 |
260012_at |
AT1G67865
|
unknown |
-0.504 |
0.039 |
| 142 |
262373_at |
AT1G73120
|
unknown |
-0.295 |
-0.060 |
| 143 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.275 |
0.135 |
| 144 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.265 |
0.160 |
| 145 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.263 |
-0.014 |
| 146 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.255 |
0.018 |
| 147 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.246 |
0.126 |
| 148 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.242 |
0.062 |
| 149 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.241 |
0.061 |
| 150 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
-0.240 |
0.050 |
| 151 |
258392_at |
AT3G15400
|
ATA20, anther 20 |
-0.232 |
-0.089 |
| 152 |
252574_at |
AT3G45430
|
LecRK-I.5, L-type lectin receptor kinase I.5 |
-0.226 |
-0.052 |
| 153 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.225 |
0.065 |
| 154 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.216 |
-0.239 |
| 155 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.213 |
0.072 |
| 156 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.210 |
-0.143 |
| 157 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.209 |
-0.348 |
| 158 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.202 |
0.055 |
| 159 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.200 |
-0.019 |
| 160 |
251483_at |
AT3G59650
|
[mitochondrial ribosomal protein L51/S25/CI-B8 family protein] |
-0.198 |
-0.043 |
| 161 |
257322_at |
ATMG01180
|
ORF111B |
-0.194 |
0.245 |
| 162 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.192 |
0.024 |
| 163 |
261203_at |
AT1G12845
|
unknown |
-0.190 |
-0.002 |
| 164 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
-0.190 |
0.051 |
| 165 |
261898_at |
AT1G80720
|
[Mitochondrial glycoprotein family protein] |
-0.187 |
-0.061 |
| 166 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
-0.185 |
-0.044 |
| 167 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.184 |
-0.072 |
| 168 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.182 |
-0.042 |
| 169 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
-0.181 |
0.203 |
| 170 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.181 |
0.141 |
| 171 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.177 |
0.030 |
| 172 |
247800_at |
AT5G58570
|
unknown |
-0.177 |
-0.196 |
| 173 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.177 |
0.085 |
| 174 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
-0.173 |
-0.097 |
| 175 |
257054_at |
AT3G15353
|
ATMT3, MT3, metallothionein 3 |
-0.173 |
0.061 |
| 176 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.168 |
0.176 |
| 177 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
-0.166 |
0.010 |
| 178 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.165 |
-0.006 |
| 179 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.164 |
-0.185 |
| 180 |
256715_at |
AT2G34090
|
MEE18, maternal effect embryo arrest 18 |
-0.163 |
0.007 |
| 181 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.162 |
-0.052 |
| 182 |
256906_at |
AT3G24000
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.161 |
-0.029 |
| 183 |
247349_at |
AT5G63820
|
unknown |
-0.159 |
-0.055 |
| 184 |
259661_at |
AT1G55265
|
unknown |
-0.158 |
0.118 |
| 185 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.155 |
0.001 |
| 186 |
250198_at |
AT5G14100
|
ABCI11, ATP-binding cassette I11, ATNAP14, ARABIDOPSIS THALIANANON-INTRINSIC ABC PROTEIN 14, NAP14, non-intrinsic ABC protein 14 |
-0.155 |
0.067 |
| 187 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.155 |
0.123 |
| 188 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
-0.151 |
-0.025 |
| 189 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.151 |
0.224 |
| 190 |
261310_at |
AT1G05750
|
CLB19, PDE247, pigment defective 247 |
-0.151 |
-0.013 |
| 191 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.150 |
0.025 |
| 192 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
-0.150 |
0.004 |
| 193 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
-0.148 |
0.075 |
| 194 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
-0.147 |
-0.071 |
| 195 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
-0.147 |
-0.157 |
| 196 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
-0.147 |
0.045 |
| 197 |
261745_at |
AT1G08500
|
AtENODL18, ENODL18, early nodulin-like protein 18 |
-0.146 |
-0.026 |
| 198 |
262920_at |
AT1G79500
|
AtkdsA1 |
-0.146 |
0.066 |
| 199 |
248704_at |
AT5G48450
|
sks3, SKU5 similar 3 |
-0.146 |
0.069 |
| 200 |
249470_at |
AT5G39350
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.145 |
-0.064 |
| 201 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.144 |
-0.048 |
| 202 |
251029_at |
AT5G02050
|
[Mitochondrial glycoprotein family protein] |
-0.144 |
-0.039 |
| 203 |
245147_at |
AT2G45280
|
ATRAD51C, RAS associated with diabetes protein 51C, RAD51C, RAS associated with diabetes protein 51C |
-0.141 |
0.024 |
| 204 |
256063_at |
AT1G07130
|
ATSTN1, STN1 |
-0.141 |
-0.016 |
| 205 |
264012_at |
AT2G21080
|
unknown |
-0.141 |
-0.018 |
| 206 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
-0.141 |
-0.048 |
| 207 |
253857_at |
AT4G27990
|
ATYLMG1-2, YLMG1-2 |
-0.141 |
0.016 |
| 208 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.140 |
0.004 |
| 209 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
-0.140 |
0.043 |
| 210 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
-0.140 |
-0.019 |
| 211 |
262892_at |
AT1G79440
|
ALDH5F1, aldehyde dehydrogenase 5F1, ENF1, ENLARGED FIL EXPRESSION DOMAIN 1, SSADH, SUCCINIC SEMIALDEHYDE DEHYDROGENASE, SSADH1, SUCCINIC SEMIALDEHYDE DEHYDROGENASE 1 |
-0.139 |
-0.046 |
| 212 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.139 |
-0.031 |
| 213 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.138 |
-0.188 |
| 214 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
-0.138 |
-0.058 |
| 215 |
260411_at |
AT1G69890
|
unknown |
-0.137 |
0.108 |
| 216 |
250148_at |
AT5G14530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.135 |
0.007 |
| 217 |
248762_at |
AT5G47455
|
unknown |
-0.135 |
0.031 |
| 218 |
259690_at |
AT1G63160
|
EMB2811, EMBRYO DEFECTIVE 2811, RFC2, replication factor C 2 |
-0.134 |
-0.055 |
| 219 |
260669_at |
AT1G19340
|
[Methyltransferase MT-A70 family protein] |
-0.134 |
0.134 |
| 220 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.134 |
-0.265 |
| 221 |
254683_at |
AT4G13800
|
unknown |
-0.134 |
0.059 |
| 222 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.133 |
0.047 |
| 223 |
260472_at |
AT1G10990
|
unknown |
-0.133 |
0.160 |
| 224 |
250127_at |
AT5G16380
|
unknown |
-0.132 |
-0.052 |
| 225 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.132 |
0.027 |
| 226 |
267639_at |
AT2G42200
|
AtSPL9, SPL9, squamosa promoter binding protein-like 9 |
-0.132 |
-0.043 |
| 227 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.131 |
-0.154 |
| 228 |
256700_at |
AT3G52260
|
[Pseudouridine synthase family protein] |
-0.131 |
0.080 |
| 229 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
-0.131 |
-0.036 |
| 230 |
261201_at |
AT1G12850
|
[Phosphoglycerate mutase family protein] |
-0.131 |
-0.028 |
| 231 |
245680_at |
AT1G56570
|
PGN, PENTATRICOPEPTIDE REPEAT PROTEIN FOR GERMINATION ON NaCl |
-0.130 |
-0.035 |
| 232 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
-0.130 |
-0.004 |
| 233 |
246393_at |
AT1G58150
|
unknown |
-0.130 |
-0.159 |
| 234 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.130 |
0.004 |
| 235 |
256269_at |
AT3G12250
|
BZIP45, TGA6, TGACG motif-binding factor 6 |
-0.128 |
0.068 |
| 236 |
247015_at |
AT5G66960
|
[Prolyl oligopeptidase family protein] |
-0.128 |
0.058 |
| 237 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.128 |
-0.104 |
| 238 |
264998_at |
AT1G67330
|
unknown |
-0.128 |
0.150 |
| 239 |
264941_at |
AT1G60680
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.127 |
-0.336 |
| 240 |
257131_at |
AT3G20240
|
[Mitochondrial substrate carrier family protein] |
-0.127 |
-0.122 |
| 241 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.127 |
0.183 |
| 242 |
266455_at |
AT2G22760
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.127 |
-0.098 |
| 243 |
257060_at |
AT3G18230
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.125 |
-0.041 |
| 244 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.125 |
-0.038 |
| 245 |
251058_at |
AT5G01790
|
unknown |
-0.124 |
-0.049 |
| 246 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.124 |
-0.072 |
| 247 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
-0.124 |
0.015 |
| 248 |
260952_at |
AT1G06140
|
MEF3, mitochondrial editing factor 3 |
-0.124 |
0.014 |
| 249 |
265790_at |
AT2G01170
|
AtGABP, BAT1, bidirectional amino acid transporter 1, GABP, GABA permease |
-0.123 |
-0.071 |
| 250 |
245600_at |
AT4G14230
|
unknown |
-0.122 |
-0.091 |
| 251 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.121 |
-0.147 |
| 252 |
246838_at |
AT5G26680
|
[5'-3' exonuclease family protein] |
-0.121 |
0.030 |
| 253 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.121 |
-0.097 |
| 254 |
249641_at |
AT5G36950
|
DEG10, degradation of periplasmic proteins 10, DegP10, DegP protease 10 |
-0.121 |
-0.050 |
| 255 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.120 |
-0.050 |
| 256 |
255791_at |
AT2G33430
|
DAL, DIFFERENTIATION AND GREENING-LIKE, DAL1, differentiation and greening-like 1, MORF2, multiple organellar RNA editing factor 2 |
-0.120 |
-0.064 |
| 257 |
245502_at |
AT4G15640
|
unknown |
-0.120 |
0.030 |
| 258 |
258990_at |
AT3G08840
|
[D-alanine--D-alanine ligase family] |
-0.120 |
0.111 |
| 259 |
257267_at |
AT3G15030
|
MEE35, maternal effect embryo arrest 35, TCP4, TCP family transcription factor 4 |
-0.119 |
-0.093 |
| 260 |
267079_at |
AT2G41200
|
unknown |
-0.119 |
0.048 |
| 261 |
261369_at |
AT1G53060
|
[Legume lectin family protein] |
-0.119 |
0.097 |
| 262 |
259943_at |
AT1G71480
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.119 |
0.085 |
| 263 |
251730_at |
AT3G56330
|
[N2,N2-dimethylguanosine tRNA methyltransferase] |
-0.119 |
-0.036 |
| 264 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.119 |
-0.059 |
| 265 |
258928_at |
AT3G10070
|
TAF12, TBP-associated factor 12, TAFII58, TATA-ASSOCIATED FACTOR II 58 |
-0.119 |
0.052 |
| 266 |
246140_at |
AT5G19910
|
MED31 |
-0.118 |
0.026 |
| 267 |
257164_at |
AT3G24320
|
ATMSH1, CHM, CHLOROPLAST MUTATOR, CHM1, MSH1, MUTL protein homolog 1 |
-0.118 |
-0.040 |
| 268 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.118 |
0.000 |
| 269 |
261508_at |
AT1G71730
|
unknown |
-0.117 |
0.019 |
| 270 |
265663_at |
AT2G24290
|
unknown |
-0.117 |
0.009 |
| 271 |
247583_at |
AT5G60750
|
SCO4, snowy cotyledon 4 |
-0.117 |
-0.007 |
| 272 |
262625_at |
AT1G06440
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.116 |
-0.052 |
| 273 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
-0.116 |
0.017 |
| 274 |
261908_at |
AT1G50650
|
[Stigma-specific Stig1 family protein] |
-0.116 |
-0.011 |
| 275 |
252509_at |
AT3G46240
|
unknown |
-0.116 |
-0.032 |