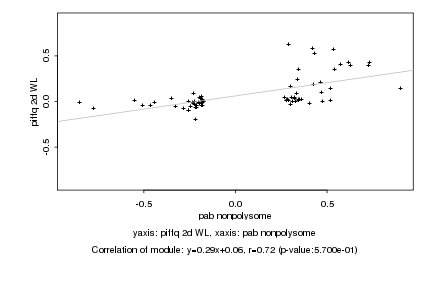
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pab nonpolysome |
Log2 signal ratio
piffq 2d WL |
| 1 |
250468_at |
AT5G10120
|
[Ethylene insensitive 3 family protein] |
0.899 |
0.151 |
| 2 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.731 |
0.432 |
| 3 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
0.722 |
0.401 |
| 4 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
0.625 |
0.398 |
| 5 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.616 |
0.431 |
| 6 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
0.573 |
0.412 |
| 7 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.539 |
0.352 |
| 8 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
0.533 |
0.577 |
| 9 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
0.518 |
0.018 |
| 10 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.515 |
0.147 |
| 11 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
0.474 |
0.003 |
| 12 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.467 |
0.109 |
| 13 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
0.461 |
0.212 |
| 14 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
0.431 |
0.532 |
| 15 |
262719_at |
AT1G43590
|
unknown |
0.423 |
0.197 |
| 16 |
250624_at |
AT5G07330
|
unknown |
0.419 |
0.587 |
| 17 |
257057_at |
AT3G15310
|
unknown |
0.402 |
-0.021 |
| 18 |
249127_at |
AT5G43500
|
ARP9, actin-related protein 9, ATARP9, actin-related protein 9 |
0.359 |
0.025 |
| 19 |
257730_at |
AT3G18420
|
[Protein prenylyltransferase superfamily protein] |
0.348 |
0.024 |
| 20 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.342 |
0.352 |
| 21 |
258134_at |
AT3G24530
|
[AAA-type ATPase family protein / ankyrin repeat family protein] |
0.339 |
0.028 |
| 22 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
0.339 |
0.016 |
| 23 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.334 |
0.245 |
| 24 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.332 |
0.093 |
| 25 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.325 |
0.008 |
| 26 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.318 |
0.052 |
| 27 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
0.317 |
0.036 |
| 28 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
0.310 |
0.010 |
| 29 |
248889_at |
AT5G46230
|
unknown |
0.302 |
0.054 |
| 30 |
247256_at |
AT5G64730
|
[Transducin/WD40 repeat-like superfamily protein] |
0.299 |
-0.032 |
| 31 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.298 |
0.168 |
| 32 |
261487_at |
AT1G14340
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.287 |
0.014 |
| 33 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
0.285 |
0.630 |
| 34 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.282 |
0.025 |
| 35 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
0.278 |
0.016 |
| 36 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.274 |
0.019 |
| 37 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.264 |
0.045 |
| 38 |
253280_at |
AT4G34110
|
ATPAB2, ARABIDOPSIS POLY(A) BINDING 2, PAB2, poly(A) binding protein 2, PABP2, POLY(A) BINDING PROTEIN 2 |
-0.855 |
-0.007 |
| 39 |
260012_at |
AT1G67865
|
unknown |
-0.780 |
-0.072 |
| 40 |
261614_at |
AT1G49760
|
PAB8, poly(A) binding protein 8, PABP8, POLY(A) BINDING PROTEIN 8 |
-0.553 |
0.015 |
| 41 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.512 |
-0.036 |
| 42 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
-0.464 |
-0.040 |
| 43 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
-0.447 |
-0.004 |
| 44 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.351 |
0.043 |
| 45 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
-0.330 |
-0.045 |
| 46 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.289 |
-0.075 |
| 47 |
258097_at |
AT3G23660
|
[Sec23/Sec24 protein transport family protein] |
-0.260 |
0.010 |
| 48 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.260 |
-0.095 |
| 49 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.258 |
-0.092 |
| 50 |
250136_at |
AT5G15380
|
DRM1, domains rearranged methylase 1 |
-0.248 |
-0.048 |
| 51 |
245085_at |
AT2G23350
|
PAB4, poly(A) binding protein 4, PABP4, POLY(A) BINDING PROTEIN 4 |
-0.239 |
-0.009 |
| 52 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.232 |
0.092 |
| 53 |
249621_at |
AT5G37530
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.232 |
-0.015 |
| 54 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.228 |
0.010 |
| 55 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.226 |
-0.027 |
| 56 |
260004_at |
AT1G67860
|
unknown |
-0.220 |
-0.064 |
| 57 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.219 |
-0.196 |
| 58 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
-0.216 |
-0.041 |
| 59 |
254169_at |
AT4G24290
|
[MAC/Perforin domain-containing protein] |
-0.214 |
-0.056 |
| 60 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.214 |
-0.039 |
| 61 |
259781_at |
AT1G29650
|
[non-LTR retrotransposon family (LINE), has a 1.5e-28 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-0.207 |
-0.010 |
| 62 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.199 |
0.049 |
| 63 |
260936_at |
AT1G45150
|
unknown |
-0.198 |
-0.017 |
| 64 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
-0.197 |
-0.019 |
| 65 |
266064_at |
AT2G18780
|
[F-box and associated interaction domains-containing protein] |
-0.196 |
0.040 |
| 66 |
255801_at |
AT4G10130
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.188 |
-0.021 |
| 67 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
-0.188 |
0.063 |
| 68 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
-0.187 |
-0.037 |
| 69 |
256389_at |
AT3G06220
|
[AP2/B3-like transcriptional factor family protein] |
-0.185 |
0.025 |
| 70 |
259569_at |
AT1G20480
|
[AMP-dependent synthetase and ligase family protein] |
-0.184 |
0.004 |
| 71 |
260669_at |
AT1G19340
|
[Methyltransferase MT-A70 family protein] |
-0.182 |
0.013 |
| 72 |
259514_at |
AT1G12480
|
CDI3, CARBON DIOXIDE INSENSITIVE 3, OZS1, OZONE-SENSITIVE 1, RCD3, RADICAL-INDUCED CELL DEATH 3, SLAC1, SLOW ANION CHANNEL-ASSOCIATED 1 |
-0.181 |
-0.040 |
| 73 |
248554_at |
AT5G50330
|
[Protein kinase superfamily protein] |
-0.181 |
-0.007 |
| 74 |
258261_at |
AT3G15740
|
[RING/U-box superfamily protein] |
-0.177 |
0.005 |