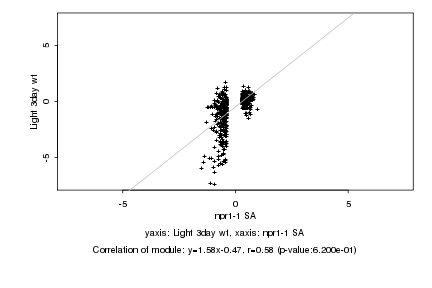
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
npr1-1 SA |
Log2 signal ratio
Light 3day wt |
| 1 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
0.941 |
-0.674 |
| 2 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
0.803 |
0.707 |
| 3 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.794 |
0.361 |
| 4 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
0.793 |
0.257 |
| 5 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
0.751 |
0.790 |
| 6 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
0.749 |
0.103 |
| 7 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
0.737 |
0.136 |
| 8 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
0.732 |
0.327 |
| 9 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
0.723 |
0.466 |
| 10 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.718 |
0.777 |
| 11 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
0.694 |
0.230 |
| 12 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
0.688 |
-0.347 |
| 13 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.686 |
0.706 |
| 14 |
256675_at |
AT3G52170
|
[DNA binding] |
0.681 |
0.545 |
| 15 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
0.676 |
0.452 |
| 16 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.674 |
0.411 |
| 17 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
0.672 |
0.105 |
| 18 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.664 |
-0.631 |
| 19 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
0.637 |
0.753 |
| 20 |
247474_at |
AT5G62280
|
unknown |
0.636 |
-1.135 |
| 21 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.635 |
0.831 |
| 22 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.634 |
-0.472 |
| 23 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
0.628 |
0.242 |
| 24 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.620 |
-0.937 |
| 25 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
0.620 |
0.303 |
| 26 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
0.613 |
0.859 |
| 27 |
262598_at |
AT1G15260
|
unknown |
0.610 |
0.441 |
| 28 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.605 |
0.897 |
| 29 |
258708_at |
AT3G09580
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.602 |
0.202 |
| 30 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
0.601 |
0.895 |
| 31 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
0.600 |
0.406 |
| 32 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.593 |
0.286 |
| 33 |
258327_at |
AT3G22640
|
PAP85 |
0.589 |
-0.515 |
| 34 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.585 |
-0.166 |
| 35 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
0.585 |
0.032 |
| 36 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
0.582 |
0.045 |
| 37 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
0.581 |
-0.592 |
| 38 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
0.577 |
1.283 |
| 39 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.572 |
-0.211 |
| 40 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.567 |
0.069 |
| 41 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
0.560 |
0.053 |
| 42 |
246908_at |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
0.558 |
0.972 |
| 43 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
0.554 |
0.526 |
| 44 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
0.552 |
0.400 |
| 45 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.537 |
-1.481 |
| 46 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
0.536 |
-0.050 |
| 47 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
0.533 |
0.277 |
| 48 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
0.527 |
0.531 |
| 49 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
0.524 |
-0.087 |
| 50 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
0.522 |
-0.452 |
| 51 |
260683_at |
AT1G17560
|
HLL, HUELLENLOS |
0.521 |
-0.081 |
| 52 |
250102_at |
AT5G16590
|
LRR1, Leucine rich repeat protein 1 |
0.520 |
0.308 |
| 53 |
250189_at |
AT5G14410
|
unknown |
0.517 |
0.776 |
| 54 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
0.516 |
0.303 |
| 55 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
0.514 |
0.417 |
| 56 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
0.511 |
0.505 |
| 57 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.510 |
-0.240 |
| 58 |
250528_at |
AT5G08600
|
[U3 ribonucleoprotein (Utp) family protein] |
0.510 |
0.259 |
| 59 |
250327_at |
AT5G12050
|
unknown |
0.509 |
0.052 |
| 60 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
0.506 |
0.258 |
| 61 |
252971_at |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
0.506 |
0.549 |
| 62 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
0.504 |
0.485 |
| 63 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.503 |
0.401 |
| 64 |
262181_at |
AT1G78060
|
[Glycosyl hydrolase family protein] |
0.503 |
0.311 |
| 65 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
0.502 |
-0.562 |
| 66 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
0.501 |
0.565 |
| 67 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.494 |
-0.515 |
| 68 |
260685_at |
AT1G17650
|
AtGLYR2, GLYR2, glyoxylate reductase 2, GR2, GLYOXYLATE REDUCTASE 2 |
0.494 |
0.109 |
| 69 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
0.494 |
0.234 |
| 70 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
0.492 |
0.535 |
| 71 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
0.488 |
-0.515 |
| 72 |
258289_at |
AT3G23450
|
unknown |
0.486 |
0.837 |
| 73 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
0.481 |
0.426 |
| 74 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.480 |
0.403 |
| 75 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.474 |
-0.573 |
| 76 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
0.471 |
0.393 |
| 77 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
0.470 |
0.500 |
| 78 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
0.465 |
0.225 |
| 79 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
0.464 |
-1.055 |
| 80 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
0.462 |
0.536 |
| 81 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.460 |
-0.112 |
| 82 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
0.458 |
0.105 |
| 83 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
0.458 |
0.172 |
| 84 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
0.456 |
-1.238 |
| 85 |
258919_at |
AT3G10525
|
LGO, LOSS OF GIANT CELLS FROM ORGANS, SMR1, SIAMESE RELATED 1 |
0.456 |
0.574 |
| 86 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
0.456 |
-0.159 |
| 87 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
0.455 |
0.383 |
| 88 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
0.455 |
-0.052 |
| 89 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.453 |
-0.183 |
| 90 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
0.449 |
0.223 |
| 91 |
256288_at |
AT3G12270
|
ATPRMT3, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 3, PRMT3, protein arginine methyltransferase 3 |
0.449 |
0.297 |
| 92 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
0.444 |
0.028 |
| 93 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
0.442 |
0.218 |
| 94 |
265326_at |
AT2G18220
|
[Noc2p family] |
0.442 |
0.469 |
| 95 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.441 |
0.766 |
| 96 |
262373_at |
AT1G73120
|
unknown |
0.441 |
-1.055 |
| 97 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
0.441 |
0.179 |
| 98 |
264673_at |
AT1G09795
|
ATATP-PRT2, ATP phosphoribosyl transferase 2, ATP-PRT2, ATP phosphoribosyl transferase 2, HISN1B |
0.440 |
0.129 |
| 99 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
0.440 |
1.043 |
| 100 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
0.439 |
0.085 |
| 101 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
0.439 |
0.442 |
| 102 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
0.436 |
-0.603 |
| 103 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.434 |
1.027 |
| 104 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.434 |
-0.397 |
| 105 |
258782_at |
AT3G11750
|
FOLB1 |
0.434 |
0.121 |
| 106 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
0.433 |
0.145 |
| 107 |
263039_at |
AT1G23280
|
[MAK16 protein-related] |
0.433 |
0.390 |
| 108 |
247339_at |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
0.431 |
0.408 |
| 109 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
0.431 |
0.677 |
| 110 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
0.430 |
0.224 |
| 111 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
0.426 |
-0.366 |
| 112 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.426 |
0.344 |
| 113 |
248854_at |
AT5G46580
|
[pentatricopeptide (PPR) repeat-containing protein] |
0.425 |
0.244 |
| 114 |
260425_at |
AT1G72440
|
EDA25, embryo sac development arrest 25, SWA2, SLOW WALKER2 |
0.425 |
0.205 |
| 115 |
256544_at |
AT1G42560
|
ATMLO9, ARABIDOPSIS THALIANA MILDEW RESISTANCE LOCUS O 9, MLO9, MILDEW RESISTANCE LOCUS O 9 |
0.424 |
-0.541 |
| 116 |
255104_at |
AT4G08685
|
SAH7 |
0.424 |
0.148 |
| 117 |
247826_at |
AT5G58480
|
[O-Glycosyl hydrolases family 17 protein] |
0.422 |
0.299 |
| 118 |
265831_at |
AT2G14460
|
unknown |
0.419 |
0.611 |
| 119 |
263474_at |
AT2G31725
|
unknown |
0.414 |
-0.054 |
| 120 |
258079_at |
AT3G25940
|
[TFIIB zinc-binding protein] |
0.413 |
0.451 |
| 121 |
247903_at |
AT5G57340
|
unknown |
0.411 |
0.552 |
| 122 |
262753_at |
AT1G16340
|
ATKDSA2, ATKSDA |
0.411 |
0.298 |
| 123 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
0.409 |
0.198 |
| 124 |
263709_at |
AT1G09310
|
unknown |
0.407 |
0.647 |
| 125 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
0.405 |
-0.174 |
| 126 |
258196_at |
AT3G13980
|
unknown |
0.404 |
-0.039 |
| 127 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
0.404 |
0.504 |
| 128 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
0.403 |
0.070 |
| 129 |
258369_at |
AT3G14310
|
ATPME3, pectin methylesterase 3, OZS2, OVERLY ZINC SENSITIVE 2, PME3, pectin methylesterase 3 |
0.402 |
-0.311 |
| 130 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
0.401 |
0.368 |
| 131 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
0.400 |
0.076 |
| 132 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.398 |
-0.021 |
| 133 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.397 |
0.173 |
| 134 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.396 |
0.351 |
| 135 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
0.395 |
0.838 |
| 136 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
0.394 |
0.142 |
| 137 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
0.394 |
-0.227 |
| 138 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
0.388 |
0.690 |
| 139 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.388 |
0.301 |
| 140 |
260565_at |
AT2G43800
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.388 |
0.721 |
| 141 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
0.388 |
0.449 |
| 142 |
254034_at |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
0.387 |
0.507 |
| 143 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.385 |
0.626 |
| 144 |
252659_at |
AT3G44430
|
unknown |
0.385 |
0.609 |
| 145 |
245712_at |
AT5G04360
|
ATLDA, limit dextrinase, ATPU1, PULLULANASE 1, LDA, limit dextrinase, PU1, PULLULANASE 1 |
0.383 |
0.310 |
| 146 |
266293_at |
AT2G29360
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.382 |
0.429 |
| 147 |
251183_at |
AT3G62630
|
unknown |
0.381 |
0.453 |
| 148 |
261387_at |
AT1G05410
|
unknown |
0.381 |
0.201 |
| 149 |
248923_at |
AT5G45940
|
AtNUDT11, Arabidopsis thaliana nudix hydrolase homolog 11, AtNUDX11, Arabidopsis thaliana nudix hydrolase homolog 11, NUDT11, nudix hydrolase homolog 11, NUDX11, nudix hydrolase homolog 11 |
0.381 |
0.793 |
| 150 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.381 |
0.161 |
| 151 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
0.381 |
0.575 |
| 152 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
0.380 |
-0.248 |
| 153 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
0.380 |
0.116 |
| 154 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
0.378 |
0.424 |
| 155 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
0.376 |
0.093 |
| 156 |
262980_at |
AT1G75680
|
AtGH9B7, glycosyl hydrolase 9B7, GH9B7, glycosyl hydrolase 9B7 |
0.372 |
0.476 |
| 157 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.372 |
-0.397 |
| 158 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
0.371 |
0.301 |
| 159 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.371 |
0.100 |
| 160 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.370 |
0.401 |
| 161 |
264096_at |
AT1G78995
|
unknown |
0.370 |
0.567 |
| 162 |
265596_at |
AT2G20020
|
ATCAF1, CAF1 |
0.370 |
0.026 |
| 163 |
247486_at |
AT5G62140
|
unknown |
0.369 |
0.755 |
| 164 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.369 |
0.657 |
| 165 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
0.368 |
0.586 |
| 166 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.368 |
0.426 |
| 167 |
247098_at |
AT5G66470
|
[RNA binding] |
0.367 |
0.070 |
| 168 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
0.366 |
0.358 |
| 169 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
0.366 |
0.073 |
| 170 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
0.365 |
-0.344 |
| 171 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.365 |
0.703 |
| 172 |
259505_at |
AT1G15810
|
[S15/NS1, RNA-binding protein] |
0.365 |
0.102 |
| 173 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
0.363 |
0.084 |
| 174 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
0.363 |
0.279 |
| 175 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.362 |
-0.600 |
| 176 |
267606_at |
AT2G26640
|
KCS11, 3-ketoacyl-CoA synthase 11 |
0.362 |
0.198 |
| 177 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.361 |
-0.479 |
| 178 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
0.361 |
0.295 |
| 179 |
260548_at |
AT2G43360
|
BIO2, BIOTIN AUXOTROPH 2, BIOB, BIOTIN AUXOTROPH B |
0.360 |
0.318 |
| 180 |
246487_at |
AT5G16030
|
unknown |
0.357 |
0.623 |
| 181 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.356 |
-0.136 |
| 182 |
266237_at |
AT2G29540
|
ATRPAC14, ATRPC14, RNApolymerase 14 kDa subunit, RPC14, RNApolymerase 14 kDa subunit |
0.356 |
0.242 |
| 183 |
263846_at |
AT2G36990
|
ATSIG6, SIGMA FACTOR 6, SIG6, SIGMA FACTOR 6, SIGF, RNApolymerase sigma-subunit F, SOLDAT8 |
0.356 |
0.314 |
| 184 |
259021_at |
AT3G07540
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.355 |
-0.149 |
| 185 |
247754_at |
AT5G59080
|
unknown |
0.355 |
0.169 |
| 186 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.354 |
-0.515 |
| 187 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
0.354 |
0.387 |
| 188 |
259422_at |
AT1G13810
|
[Restriction endonuclease, type II-like superfamily protein] |
0.353 |
0.574 |
| 189 |
258432_at |
AT3G16570
|
ATRALF23, ARABIDOPSIS RAPID ALKALINIZATION FACTOR 23, RALF23, rapid alkalinization factor 23 |
0.352 |
0.293 |
| 190 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
0.352 |
1.379 |
| 191 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
0.352 |
-0.515 |
| 192 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
0.350 |
0.019 |
| 193 |
260368_at |
AT1G69700
|
ATHVA22C, HVA22 homologue C, HVA22C, HVA22 homologue C |
0.349 |
0.290 |
| 194 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
0.349 |
0.172 |
| 195 |
251788_at |
AT3G55420
|
unknown |
0.347 |
0.450 |
| 196 |
253028_at |
AT4G38160
|
pde191, pigment defective 191 |
0.347 |
0.230 |
| 197 |
257974_at |
AT3G20820
|
[Leucine-rich repeat (LRR) family protein] |
0.346 |
0.341 |
| 198 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
0.346 |
0.260 |
| 199 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
0.346 |
0.156 |
| 200 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
0.345 |
0.090 |
| 201 |
266189_at |
AT2G39020
|
NATA2 |
0.345 |
0.159 |
| 202 |
259431_at |
AT1G01620
|
PIP1;3, PLASMA MEMBRANE INTRINSIC PROTEIN 1;3, PIP1C, plasma membrane intrinsic protein 1C, TMP-B |
0.344 |
-0.080 |
| 203 |
264963_at |
AT1G60600
|
ABC4, ABERRANT CHLOROPLAST DEVELOPMENT 4 |
0.344 |
0.444 |
| 204 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
0.343 |
-0.062 |
| 205 |
267192_at |
AT2G30890
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
0.343 |
-0.139 |
| 206 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.342 |
0.137 |
| 207 |
251864_at |
AT3G54920
|
PMR6, powdery mildew resistant 6 |
0.341 |
0.395 |
| 208 |
267586_at |
AT2G41950
|
unknown |
0.340 |
0.289 |
| 209 |
252115_at |
AT3G51600
|
LTP5, lipid transfer protein 5 |
0.340 |
-0.022 |
| 210 |
259788_at |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.339 |
0.529 |
| 211 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.338 |
0.323 |
| 212 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
0.337 |
0.305 |
| 213 |
258054_at |
AT3G16240
|
AQP1, ATTIP2;1, DELTA-TIP, delta tonoplast integral protein, DELTA-TIP1, TIP2;1 |
0.336 |
-0.061 |
| 214 |
248230_at |
AT5G53830
|
[VQ motif-containing protein] |
0.336 |
-0.594 |
| 215 |
248409_at |
AT5G51545
|
LPA2, low psii accumulation2 |
0.335 |
0.101 |
| 216 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
0.335 |
0.063 |
| 217 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.334 |
-0.183 |
| 218 |
248402_at |
AT5G52100
|
CRR1, chlororespiration reduction 1 |
0.334 |
0.184 |
| 219 |
254225_at |
AT4G23670
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.333 |
0.376 |
| 220 |
259818_at |
AT1G49890
|
QWRF2, QWRF domain containing 2 |
0.331 |
0.260 |
| 221 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
0.331 |
-0.152 |
| 222 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.331 |
0.320 |
| 223 |
246576_at |
AT1G31650
|
ATROPGEF14, ROPGEF14, ROP (rho of plants) guanine nucleotide exchange factor 14 |
0.330 |
0.308 |
| 224 |
254649_at |
AT4G18570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.330 |
0.406 |
| 225 |
266303_at |
AT2G27060
|
[Leucine-rich repeat protein kinase family protein] |
0.327 |
0.160 |
| 226 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
0.326 |
-0.252 |
| 227 |
263326_at |
AT2G04280
|
unknown |
0.326 |
0.259 |
| 228 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
0.325 |
0.126 |
| 229 |
262573_at |
AT1G15390
|
ATDEF1, PDF1A, peptide deformylase 1A |
0.325 |
0.034 |
| 230 |
248261_at |
AT5G53280
|
PDV1, PLASTID DIVISION1 |
0.323 |
0.600 |
| 231 |
253233_at |
AT4G34290
|
[SWIB/MDM2 domain superfamily protein] |
0.323 |
-0.002 |
| 232 |
260632_at |
AT1G62360
|
BUM, BUMBERSHOOT, BUM1, BUMBERSHOOT 1, SHL, SHOOTLESS, STM, SHOOT MERISTEMLESS, WAM, WALDMEISTER, WAM1, WALDMEISTER 1 |
0.323 |
-0.515 |
| 233 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
0.323 |
0.263 |
| 234 |
263496_at |
AT2G42570
|
TBL39, TRICHOME BIREFRINGENCE-LIKE 39 |
0.323 |
0.689 |
| 235 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
0.322 |
0.498 |
| 236 |
267310_at |
AT2G34680
|
AIR9, AUXIN-INDUCED IN ROOT CULTURES 9 |
0.322 |
0.027 |
| 237 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
0.321 |
0.084 |
| 238 |
245952_at |
AT5G28500
|
unknown |
0.320 |
-0.054 |
| 239 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
0.320 |
-0.263 |
| 240 |
265873_at |
AT2G01630
|
[O-Glycosyl hydrolases family 17 protein] |
0.319 |
0.091 |
| 241 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
0.319 |
-0.417 |
| 242 |
250837_at |
AT5G04620
|
ATBIOF, biotin F, BIO4, biotin 4, BIOF, biotin F |
0.319 |
0.311 |
| 243 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
0.319 |
0.822 |
| 244 |
256796_at |
AT3G22210
|
unknown |
0.318 |
0.473 |
| 245 |
260157_at |
AT1G52930
|
[Ribosomal RNA processing Brix domain protein] |
0.317 |
0.163 |
| 246 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
0.317 |
-0.514 |
| 247 |
250884_at |
AT5G03940
|
54CP, 54 CHLOROPLAST PROTEIN, CPSRP54, chloroplast signal recognition particle 54 kDa subunit, FFC, FIFTY-FOUR CHLOROPLAST HOMOLOGUE, SRP54CP, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT CHLOROPLAST PROTEIN |
0.317 |
0.200 |
| 248 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
0.316 |
0.238 |
| 249 |
263371_at |
AT2G20490
|
EDA27, EMBRYO SAC DEVELOPMENT ARREST 27, NOP10 |
0.315 |
-0.052 |
| 250 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
0.314 |
-0.200 |
| 251 |
245743_at |
AT1G51080
|
unknown |
0.314 |
0.676 |
| 252 |
251667_at |
AT3G57150
|
AtCBF5, AtNAP57, CBF5, NAP57, homologue of NAP57 |
0.313 |
0.118 |
| 253 |
246926_at |
AT5G25240
|
unknown |
0.313 |
-0.077 |
| 254 |
265154_at |
AT1G30960
|
[GTP-binding family protein] |
0.312 |
-0.090 |
| 255 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
0.312 |
-0.591 |
| 256 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
0.311 |
0.496 |
| 257 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
0.311 |
0.179 |
| 258 |
249113_at |
AT5G43790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.310 |
0.337 |
| 259 |
255889_at |
AT1G17840
|
ABCG11, ATP-binding cassette G11, AtABCG11, ATWBC11, ARABIDOPSIS THALIANA WHITE-BROWN COMPLEX HOMOLOG PROTEIN 11, COF1, CUTICULAR DEFECT AND ORGAN FUSION 1, DSO, DESPERADO, WBC11, white-brown complex homolog protein 11 |
0.310 |
0.691 |
| 260 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.309 |
0.340 |
| 261 |
260363_at |
AT1G70550
|
unknown |
0.309 |
0.642 |
| 262 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.309 |
-0.129 |
| 263 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
0.309 |
0.191 |
| 264 |
251736_at |
AT3G56130
|
[biotin/lipoyl attachment domain-containing protein] |
0.308 |
0.168 |
| 265 |
251008_at |
AT5G02710
|
unknown |
0.308 |
0.054 |
| 266 |
256341_at |
AT1G72040
|
AtdNK, dNK, deoxyribonucleoside kinase |
0.308 |
0.197 |
| 267 |
262594_at |
AT1G15250
|
[Zinc-binding ribosomal protein family protein] |
0.308 |
0.047 |
| 268 |
260165_at |
AT1G79850
|
CS17, PDE347, PIGMENT DEFECTIVE 347, PRPS17, PLASTID RIBOSOMAL SMALL SUBUNIT PROTEIN 17, RPS17, ribosomal protein S17 |
0.308 |
-0.207 |
| 269 |
258529_at |
AT3G06740
|
GATA15, GATA transcription factor 15 |
0.305 |
0.232 |
| 270 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.305 |
0.004 |
| 271 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
0.305 |
0.392 |
| 272 |
256065_at |
AT1G07070
|
[Ribosomal protein L35Ae family protein] |
0.305 |
0.020 |
| 273 |
249554_at |
AT5G38290
|
[Peptidyl-tRNA hydrolase family protein] |
0.305 |
0.345 |
| 274 |
258156_at |
AT3G18050
|
unknown |
0.304 |
0.409 |
| 275 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
0.304 |
-0.008 |
| 276 |
260902_at |
AT1G21440
|
[Phosphoenolpyruvate carboxylase family protein] |
0.303 |
0.051 |
| 277 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.303 |
-0.232 |
| 278 |
259830_at |
AT1G80630
|
[RNI-like superfamily protein] |
0.302 |
0.043 |
| 279 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.302 |
0.185 |
| 280 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.302 |
0.038 |
| 281 |
252724_at |
AT3G43540
|
unknown |
0.302 |
0.292 |
| 282 |
263287_at |
AT2G36145
|
unknown |
0.301 |
0.100 |
| 283 |
260267_at |
AT1G68530
|
CER6, ECERIFERUM 6, CUT1, CUTICULAR 1, G2, KCS6, 3-ketoacyl-CoA synthase 6, POP1, POLLEN-PISTIL INCOMPATIBILITY 1 |
0.301 |
0.758 |
| 284 |
246097_at |
AT5G20270
|
HHP1, heptahelical transmembrane protein1 |
0.300 |
0.965 |
| 285 |
257724_at |
AT3G18510
|
unknown |
0.300 |
0.167 |
| 286 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.300 |
0.566 |
| 287 |
265895_at |
AT2G15000
|
unknown |
0.300 |
-0.074 |
| 288 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.533 |
-5.909 |
| 289 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-1.432 |
-5.395 |
| 290 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-1.377 |
-4.893 |
| 291 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-1.302 |
-1.845 |
| 292 |
260278_at |
AT1G80590
|
ATWRKY66, WRKY66, WRKY DNA-binding protein 66 |
-1.260 |
-0.515 |
| 293 |
245226_at |
AT3G29970
|
[B12D protein] |
-1.197 |
-0.515 |
| 294 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-1.172 |
-5.072 |
| 295 |
248987_at |
AT5G45180
|
[Flavin-binding monooxygenase family protein] |
-1.132 |
-0.515 |
| 296 |
264861_at |
AT1G24320
|
[Six-hairpin glycosidases superfamily protein] |
-1.120 |
-0.515 |
| 297 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
-1.117 |
-7.301 |
| 298 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.102 |
-0.422 |
| 299 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-1.093 |
-5.051 |
| 300 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-1.088 |
-2.391 |
| 301 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-1.044 |
-1.082 |
| 302 |
267572_at |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-1.038 |
-0.515 |
| 303 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-1.014 |
-5.802 |
| 304 |
249295_at |
AT5G41280
|
[Receptor-like protein kinase-related family protein] |
-0.983 |
-2.544 |
| 305 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.971 |
-6.236 |
| 306 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
-0.949 |
-4.039 |
| 307 |
258203_at |
AT3G13950
|
unknown |
-0.945 |
-5.304 |
| 308 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.944 |
-2.271 |
| 309 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.944 |
-7.304 |
| 310 |
267573_at |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.942 |
-0.515 |
| 311 |
253665_at |
AT4G30230
|
unknown |
-0.940 |
-0.101 |
| 312 |
249476_at |
AT5G38910
|
[RmlC-like cupins superfamily protein] |
-0.940 |
-0.515 |
| 313 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.939 |
0.163 |
| 314 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.919 |
-1.274 |
| 315 |
247264_at |
AT5G64530
|
ANAC104, Arabidopsis NAC domain containing protein 104, XND1, xylem NAC domain 1 |
-0.909 |
-0.215 |
| 316 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.906 |
-0.327 |
| 317 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
-0.889 |
-1.752 |
| 318 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.876 |
-1.081 |
| 319 |
257810_at |
AT3G27070
|
TOM20-1, translocase outer membrane 20-1 |
-0.875 |
-0.515 |
| 320 |
254249_at |
AT4G23280
|
CRK20, cysteine-rich RLK (RECEPTOR-like protein kinase) 20 |
-0.872 |
-3.450 |
| 321 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.867 |
-1.531 |
| 322 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
-0.865 |
-2.679 |
| 323 |
248717_at |
AT5G48175
|
unknown |
-0.860 |
0.799 |
| 324 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.853 |
-2.625 |
| 325 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-0.832 |
1.225 |
| 326 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
-0.821 |
-2.033 |
| 327 |
259260_at |
AT3G11370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.820 |
-0.515 |
| 328 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.819 |
-0.515 |
| 329 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
-0.814 |
0.006 |
| 330 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
-0.813 |
-1.055 |
| 331 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
-0.806 |
-0.704 |
| 332 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.804 |
0.147 |
| 333 |
253795_at |
AT4G28420
|
[Tyrosine transaminase family protein] |
-0.804 |
0.006 |
| 334 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.801 |
-1.268 |
| 335 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
-0.789 |
-5.675 |
| 336 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.776 |
0.524 |
| 337 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
-0.774 |
-1.938 |
| 338 |
248316_at |
AT5G52670
|
[Copper transport protein family] |
-0.770 |
-0.515 |
| 339 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
-0.768 |
-4.846 |
| 340 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.761 |
-4.392 |
| 341 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.761 |
-5.160 |
| 342 |
257924_at |
AT3G23190
|
[HR-like lesion-inducing protein-related] |
-0.760 |
-0.025 |
| 343 |
248319_at |
AT5G52710
|
[Copper transport protein family] |
-0.757 |
-1.055 |
| 344 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
-0.750 |
0.700 |
| 345 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.747 |
-3.158 |
| 346 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
-0.732 |
-2.157 |
| 347 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
-0.727 |
-1.447 |
| 348 |
252582_at |
AT3G45530
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.727 |
-0.515 |
| 349 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
-0.720 |
-0.999 |
| 350 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
-0.713 |
-2.941 |
| 351 |
251689_at |
AT3G56500
|
[serine-rich protein-related] |
-0.706 |
-2.544 |
| 352 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
-0.704 |
-2.798 |
| 353 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.700 |
-2.150 |
| 354 |
259213_at |
AT3G09010
|
[Protein kinase superfamily protein] |
-0.699 |
-0.695 |
| 355 |
246406_at |
AT1G57650
|
[ATP binding] |
-0.698 |
-3.564 |
| 356 |
254598_at |
AT4G18990
|
XTH29, xyloglucan endotransglucosylase/hydrolase 29 |
-0.697 |
-0.295 |
| 357 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
-0.697 |
-0.515 |
| 358 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.696 |
-3.504 |
| 359 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-0.695 |
-0.515 |
| 360 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
-0.691 |
-2.241 |
| 361 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.689 |
-1.802 |
| 362 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.687 |
-1.542 |
| 363 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-0.685 |
0.345 |
| 364 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.676 |
-3.766 |
| 365 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.673 |
-5.505 |
| 366 |
267207_at |
AT2G30840
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.671 |
-0.535 |
| 367 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
-0.670 |
0.386 |
| 368 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.668 |
-2.999 |
| 369 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.665 |
-2.798 |
| 370 |
254894_at |
AT4G11840
|
PLDGAMMA3, phospholipase D gamma 3 |
-0.663 |
-0.796 |
| 371 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
-0.662 |
-4.798 |
| 372 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.657 |
-1.179 |
| 373 |
257535_at |
AT3G09490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.654 |
-1.407 |
| 374 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.650 |
-2.950 |
| 375 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
-0.646 |
-3.175 |
| 376 |
249896_at |
AT5G22530
|
unknown |
-0.645 |
-3.854 |
| 377 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.645 |
-2.506 |
| 378 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
-0.645 |
-1.919 |
| 379 |
245076_at |
AT2G23170
|
GH3.3 |
-0.642 |
-1.103 |
| 380 |
254847_at |
AT4G11850
|
MEE54, maternal effect embryo arrest 54, PLDGAMMA1, phospholipase D gamma 1 |
-0.638 |
-1.142 |
| 381 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.628 |
-0.142 |
| 382 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.627 |
-0.289 |
| 383 |
247740_at |
AT5G58940
|
CRCK1, calmodulin-binding receptor-like cytoplasmic kinase 1 |
-0.624 |
-0.788 |
| 384 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
-0.623 |
-0.914 |
| 385 |
254571_at |
AT4G19370
|
unknown |
-0.623 |
-0.821 |
| 386 |
257184_at |
AT3G13090
|
ABCC6, ATP-binding cassette C6, ATMRP8, multidrug resistance-associated protein 8, MRP8, multidrug resistance-associated protein 8 |
-0.622 |
-0.839 |
| 387 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.621 |
0.627 |
| 388 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.616 |
-2.114 |
| 389 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
-0.612 |
-0.295 |
| 390 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.611 |
0.744 |
| 391 |
245932_at |
AT5G09290
|
[Inositol monophosphatase family protein] |
-0.610 |
-3.768 |
| 392 |
249055_at |
AT5G44460
|
CML43, calmodulin like 43 |
-0.608 |
-3.375 |
| 393 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
-0.606 |
-3.671 |
| 394 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.605 |
-5.592 |
| 395 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
-0.603 |
0.185 |
| 396 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.603 |
0.553 |
| 397 |
253522_at |
AT4G31290
|
AtGGCT2;2, GGCT2;2, gamma-glutamyl cyclotransferase 2;2 |
-0.601 |
0.737 |
| 398 |
248980_at |
AT5G45090
|
AtPP2-A7, phloem protein 2-A7, PP2-A7, phloem protein 2-A7 |
-0.599 |
-3.996 |
| 399 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.598 |
0.815 |
| 400 |
259473_at |
AT1G19025
|
[DNA repair metallo-beta-lactamase family protein] |
-0.598 |
0.622 |
| 401 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
-0.596 |
-1.994 |
| 402 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.596 |
0.933 |
| 403 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.594 |
-2.150 |
| 404 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.594 |
-0.515 |
| 405 |
259261_at |
AT3G11390
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.592 |
0.006 |
| 406 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
-0.587 |
-0.378 |
| 407 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.584 |
-1.445 |
| 408 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
-0.584 |
-1.055 |
| 409 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.583 |
-0.158 |
| 410 |
252222_at |
AT3G49845
|
WIH3, WINDHOSE 3 |
-0.583 |
-0.515 |
| 411 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.581 |
-0.515 |
| 412 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
-0.580 |
-0.515 |
| 413 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
-0.579 |
-0.515 |
| 414 |
259960_at |
AT1G53710
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.576 |
0.041 |
| 415 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
-0.575 |
-0.206 |
| 416 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
-0.574 |
0.257 |
| 417 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.574 |
-0.515 |
| 418 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.573 |
-4.210 |
| 419 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.571 |
-0.820 |
| 420 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
-0.568 |
-2.411 |
| 421 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
-0.567 |
-1.565 |
| 422 |
245082_at |
AT2G23270
|
unknown |
-0.566 |
-1.752 |
| 423 |
245209_at |
AT5G12340
|
unknown |
-0.566 |
-1.447 |
| 424 |
260522_x_at |
AT2G41730
|
unknown |
-0.561 |
-0.300 |
| 425 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.561 |
-0.394 |
| 426 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
-0.559 |
-0.232 |
| 427 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
-0.558 |
-4.685 |
| 428 |
265582_at |
AT2G20030
|
[RING/U-box superfamily protein] |
-0.556 |
-0.515 |
| 429 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
-0.552 |
-0.458 |
| 430 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
-0.549 |
-5.293 |
| 431 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
-0.548 |
-1.688 |
| 432 |
254597_at |
AT4G18980
|
AtS40-3, AtS40-3 |
-0.548 |
-1.095 |
| 433 |
254292_at |
AT4G23030
|
[MATE efflux family protein] |
-0.548 |
-2.258 |
| 434 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
-0.546 |
0.006 |
| 435 |
261842_at |
AT1G16160
|
WAKL5, wall associated kinase-like 5 |
-0.546 |
-0.534 |
| 436 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
-0.546 |
-3.526 |
| 437 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.543 |
-0.515 |
| 438 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-0.542 |
0.747 |
| 439 |
255075_at |
AT4G09110
|
[RING/U-box superfamily protein] |
-0.539 |
-0.515 |
| 440 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
-0.532 |
-2.617 |
| 441 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
-0.532 |
-4.542 |
| 442 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.532 |
-1.084 |
| 443 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
-0.531 |
-0.593 |
| 444 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.530 |
-4.617 |
| 445 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.530 |
-2.001 |
| 446 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
-0.521 |
-1.994 |
| 447 |
245611_at |
AT4G14390
|
[Ankyrin repeat family protein] |
-0.521 |
-2.868 |
| 448 |
261481_at |
AT1G14260
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.519 |
-1.300 |
| 449 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.519 |
-0.585 |
| 450 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
-0.518 |
-1.157 |
| 451 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
-0.514 |
0.614 |
| 452 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
-0.512 |
-1.066 |
| 453 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
-0.510 |
-0.515 |
| 454 |
260280_at |
AT1G80580
|
[Integrase-type DNA-binding superfamily protein] |
-0.507 |
-0.515 |
| 455 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
-0.505 |
-0.019 |
| 456 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
-0.503 |
-2.903 |
| 457 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.503 |
-2.938 |
| 458 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
-0.501 |
-4.244 |
| 459 |
249406_at |
AT5G40210
|
UMAMIT42, Usually multiple acids move in and out Transporters 42 |
-0.500 |
1.299 |
| 460 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.498 |
-4.261 |
| 461 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
-0.496 |
1.157 |
| 462 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
-0.495 |
-0.515 |
| 463 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.494 |
-2.659 |
| 464 |
250701_at |
AT5G06839
|
bZIP65, TGA10, TGACG (TGA) motif-binding protein 10 |
-0.494 |
-0.515 |
| 465 |
258182_at |
AT3G21500
|
DXL1, DXS-like 1, DXPS1, 1-deoxy-D-xylulose 5-phosphate synthase 1, DXS2, 1-deoxy-D-xylulose 5-phosphate (DXP) synthase 2 |
-0.490 |
-0.515 |
| 466 |
259134_at |
AT3G05390
|
unknown |
-0.488 |
-1.447 |
| 467 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
-0.485 |
-3.109 |
| 468 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.485 |
-3.611 |
| 469 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
-0.485 |
1.732 |
| 470 |
250267_at |
AT5G12930
|
unknown |
-0.483 |
-1.326 |
| 471 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
-0.482 |
-0.635 |
| 472 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.482 |
-0.099 |
| 473 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
-0.481 |
-1.067 |
| 474 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
-0.481 |
-1.481 |
| 475 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.480 |
0.031 |
| 476 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
-0.478 |
-5.156 |
| 477 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
-0.475 |
-5.165 |
| 478 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.475 |
0.648 |
| 479 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.475 |
-0.431 |
| 480 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.473 |
-2.558 |
| 481 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
-0.473 |
-0.161 |
| 482 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
-0.472 |
-0.161 |
| 483 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
-0.472 |
-0.370 |
| 484 |
250152_at |
AT5G15120
|
unknown |
-0.469 |
-0.585 |
| 485 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
-0.469 |
-2.679 |
| 486 |
245566_at |
AT4G14610
|
[pseudogene] |
-0.468 |
-0.453 |
| 487 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
-0.468 |
-1.979 |
| 488 |
260728_at |
AT1G48210
|
[Protein kinase superfamily protein] |
-0.467 |
-1.229 |
| 489 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
-0.466 |
-0.744 |
| 490 |
258228_at |
AT3G27610
|
[Nucleotidylyl transferase superfamily protein] |
-0.464 |
0.388 |
| 491 |
247794_at |
AT5G58670
|
ATPLC, ARABIDOPSIS THALIANA PHOSPHOLIPASE C, ATPLC1, phospholipase C1, PLC1, phospholipase C 1, PLC1, phospholipase C1 |
-0.464 |
0.470 |
| 492 |
264716_at |
AT1G70170
|
MMP, matrix metalloproteinase |
-0.462 |
-1.865 |
| 493 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.460 |
0.130 |
| 494 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.459 |
0.021 |
| 495 |
246082_at |
AT5G20480
|
EFR, EF-TU receptor |
-0.459 |
-0.846 |
| 496 |
249489_at |
AT5G39090
|
[HXXXD-type acyl-transferase family protein] |
-0.458 |
-0.168 |
| 497 |
254408_at |
AT4G21390
|
B120 |
-0.454 |
-0.528 |
| 498 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.453 |
-2.525 |
| 499 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.452 |
-2.294 |
| 500 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.451 |
-0.515 |
| 501 |
253993_at |
AT4G26070
|
ATMEK1, MEK1, MAP kinase/ ERK kinase 1, MKK1, MITOGEN ACTIVATED PROTEIN KINASE KINASE 1, NMAPKK |
-0.451 |
-0.686 |
| 502 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
-0.450 |
-3.796 |
| 503 |
263854_at |
AT2G04430
|
atnudt5, nudix hydrolase homolog 5, NUDT5, nudix hydrolase homolog 5 |
-0.449 |
-1.711 |
| 504 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.449 |
0.154 |
| 505 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
-0.449 |
-0.779 |
| 506 |
246584_at |
AT5G14730
|
unknown |
-0.449 |
-0.462 |
| 507 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
-0.448 |
-2.114 |
| 508 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
-0.447 |
-5.397 |
| 509 |
265945_at |
AT2G19500
|
ATCKX2, CKX2, cytokinin oxidase 2 |
-0.446 |
-0.515 |
| 510 |
251755_at |
AT3G55790
|
unknown |
-0.444 |
-0.515 |
| 511 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
-0.440 |
-1.055 |
| 512 |
257542_at |
AT3G26050
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.439 |
0.017 |
| 513 |
265479_at |
AT2G15760
|
unknown |
-0.437 |
0.156 |
| 514 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.437 |
-2.889 |
| 515 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
-0.436 |
-1.135 |
| 516 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
-0.436 |
-0.527 |
| 517 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
-0.435 |
-0.674 |
| 518 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
-0.434 |
-0.841 |
| 519 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
-0.434 |
-0.171 |
| 520 |
253768_at |
AT4G28550
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
-0.434 |
0.076 |
| 521 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
-0.432 |
-0.242 |
| 522 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
-0.430 |
-0.515 |
| 523 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
-0.430 |
1.321 |
| 524 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
-0.430 |
-0.331 |
| 525 |
250998_at |
AT5G02620
|
ANK1, ankyrin-like1, ATANK1 |
-0.428 |
0.222 |
| 526 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
-0.428 |
-2.923 |
| 527 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
-0.428 |
-2.121 |
| 528 |
247318_at |
AT5G63990
|
[Inositol monophosphatase family protein] |
-0.428 |
-1.179 |
| 529 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.426 |
0.989 |
| 530 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
-0.426 |
-2.336 |
| 531 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
-0.426 |
-0.714 |
| 532 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
-0.426 |
0.213 |
| 533 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.426 |
-0.198 |
| 534 |
266835_at |
AT2G29990
|
NDA2, alternative NAD(P)H dehydrogenase 2 |
-0.425 |
-1.214 |
| 535 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.424 |
-1.484 |
| 536 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
-0.422 |
-3.093 |
| 537 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
-0.421 |
-4.004 |
| 538 |
256617_at |
AT3G22240
|
unknown |
-0.421 |
0.270 |
| 539 |
251063_at |
AT5G01850
|
[Protein kinase superfamily protein] |
-0.420 |
-0.529 |
| 540 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.420 |
-0.152 |
| 541 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-0.420 |
0.377 |
| 542 |
248889_at |
AT5G46230
|
unknown |
-0.420 |
-1.233 |
| 543 |
252346_at |
AT3G48650
|
[pseudogene] |
-0.419 |
-2.158 |
| 544 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
-0.419 |
-0.515 |
| 545 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
-0.419 |
-3.551 |
| 546 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.419 |
-0.777 |
| 547 |
251769_at |
AT3G55950
|
ATCRR3, CCR3, CRINKLY4 related 3 |
-0.419 |
-0.188 |
| 548 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
-0.419 |
0.013 |
| 549 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
-0.418 |
-1.752 |
| 550 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
-0.418 |
-0.004 |
| 551 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
-0.417 |
-0.618 |
| 552 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
-0.416 |
-2.621 |
| 553 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
-0.415 |
-0.669 |
| 554 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
-0.413 |
-0.221 |
| 555 |
253137_at |
AT4G35500
|
[Protein kinase superfamily protein] |
-0.413 |
0.144 |
| 556 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
-0.413 |
-0.589 |
| 557 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
-0.412 |
0.446 |
| 558 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
-0.412 |
-0.553 |
| 559 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
-0.412 |
-1.116 |
| 560 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.410 |
-0.271 |
| 561 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
-0.409 |
0.147 |
| 562 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.407 |
-0.748 |
| 563 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
-0.406 |
-3.717 |
| 564 |
254532_at |
AT4G19660
|
ATNPR4, NPR4, NPR1-like protein 4 |
-0.405 |
-0.049 |
| 565 |
259370_at |
AT1G69050
|
unknown |
-0.405 |
-1.358 |
| 566 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.404 |
-0.995 |
| 567 |
256251_at |
AT3G11330
|
PIRL9, plant intracellular ras group-related LRR 9 |
-0.404 |
-0.105 |
| 568 |
264328_at |
AT1G04100
|
IAA10, indoleacetic acid-induced protein 10 |
-0.404 |
0.194 |
| 569 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.403 |
-0.812 |
| 570 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
-0.403 |
0.123 |
| 571 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.401 |
-0.024 |
| 572 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.401 |
-1.372 |
| 573 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
-0.400 |
-2.097 |