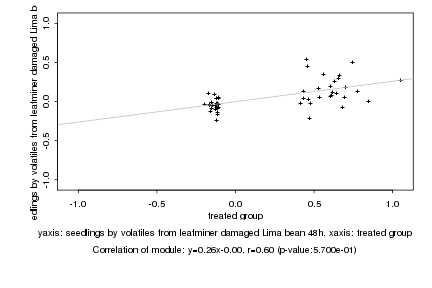
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
treated group |
Log2 signal ratio
seedlings by volatiles from leafminer damaged Lima bean 48h |
| 1 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
1.048 |
0.280 |
| 2 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.846 |
0.013 |
| 3 |
256937_at |
AT3G22620
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.776 |
0.129 |
| 4 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.745 |
0.500 |
| 5 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.698 |
0.183 |
| 6 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.689 |
0.052 |
| 7 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.676 |
-0.067 |
| 8 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.657 |
0.333 |
| 9 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
0.654 |
0.300 |
| 10 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.643 |
0.104 |
| 11 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.628 |
0.260 |
| 12 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.613 |
0.125 |
| 13 |
249454_at |
AT5G39520
|
unknown |
0.610 |
0.086 |
| 14 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.603 |
0.203 |
| 15 |
258209_at |
AT3G14060
|
unknown |
0.600 |
0.067 |
| 16 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.555 |
0.358 |
| 17 |
249289_at |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
0.530 |
0.056 |
| 18 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.526 |
0.174 |
| 19 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.473 |
-0.025 |
| 20 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.469 |
-0.211 |
| 21 |
248218_at |
AT5G53710
|
unknown |
0.460 |
0.032 |
| 22 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.456 |
0.457 |
| 23 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.446 |
0.546 |
| 24 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.427 |
0.041 |
| 25 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.427 |
0.131 |
| 26 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.414 |
-0.019 |
| 27 |
248626_at |
AT5G48940
|
[Leucine-rich repeat transmembrane protein kinase family protein] |
-0.201 |
-0.029 |
| 28 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
-0.174 |
0.106 |
| 29 |
257359_x_at |
AT2G34290
|
[Protein kinase superfamily protein] |
-0.171 |
-0.040 |
| 30 |
263733_at |
AT1G60020
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:AAC02672 polyprotein (Ty1_Copia-element) (Arabidopsis arenosa)] |
-0.169 |
-0.029 |
| 31 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.162 |
-0.120 |
| 32 |
245103_at |
AT2G41590
|
unknown |
-0.155 |
-0.005 |
| 33 |
263558_at |
AT2G16380
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.153 |
-0.088 |
| 34 |
246792_at |
AT5G27290
|
unknown |
-0.151 |
-0.043 |
| 35 |
258422_at |
AT3G16710
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.135 |
0.099 |
| 36 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.128 |
-0.100 |
| 37 |
254518_at |
AT4G19910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.127 |
-0.042 |
| 38 |
251627_at |
AT3G57270
|
BG1, beta-1,3-glucanase 1 |
-0.125 |
0.043 |
| 39 |
249111_at |
AT5G43770
|
[proline-rich family protein] |
-0.125 |
0.042 |
| 40 |
257246_at |
AT3G24130
|
[Pectin lyase-like superfamily protein] |
-0.124 |
-0.074 |
| 41 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.124 |
-0.236 |
| 42 |
257995_at |
AT3G19940
|
[Major facilitator superfamily protein] |
-0.123 |
-0.017 |
| 43 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.117 |
-0.135 |
| 44 |
254317_at |
AT4G22510
|
unknown |
-0.117 |
-0.084 |
| 45 |
254506_at |
AT4G20140
|
GSO1, GASSHO1 |
-0.116 |
-0.154 |
| 46 |
265425_at |
AT2G20770
|
GCL2, GCR2-like 2 |
-0.116 |
-0.079 |
| 47 |
266176_at |
AT2G02250
|
AtPP2-B2, phloem protein 2-B2, PP2-B2, phloem protein 2-B2 |
-0.115 |
-0.013 |
| 48 |
267398_at |
AT2G44560
|
AtGH9B11, glycosyl hydrolase 9B11, GH9B11, glycosyl hydrolase 9B11 |
-0.112 |
-0.067 |
| 49 |
246723_at |
AT5G29030
|
[pseudogene] |
-0.112 |
0.044 |
| 50 |
245820_at |
AT1G26320
|
[Zinc-binding dehydrogenase family protein] |
-0.111 |
0.052 |
| 51 |
258969_at |
AT3G10680
|
[HSP20-like chaperones superfamily protein] |
-0.108 |
-0.038 |