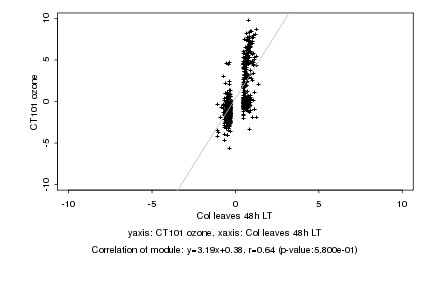
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col leaves 48h LT |
Log2 signal ratio
CT101 ozone |
| 1 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
1.360 |
2.114 |
| 2 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
1.254 |
4.344 |
| 3 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
1.212 |
5.456 |
| 4 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.210 |
-1.857 |
| 5 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
1.208 |
8.784 |
| 6 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
1.164 |
8.076 |
| 7 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
1.119 |
5.312 |
| 8 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
1.112 |
5.111 |
| 9 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
1.100 |
-0.931 |
| 10 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
1.092 |
-0.900 |
| 11 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.087 |
1.177 |
| 12 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
1.081 |
4.430 |
| 13 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
1.061 |
4.547 |
| 14 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
1.060 |
7.984 |
| 15 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
1.052 |
7.743 |
| 16 |
249896_at |
AT5G22530
|
unknown |
1.051 |
0.225 |
| 17 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
1.045 |
4.703 |
| 18 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
1.040 |
3.436 |
| 19 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
1.023 |
5.901 |
| 20 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
1.021 |
7.334 |
| 21 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
1.020 |
7.752 |
| 22 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
1.015 |
5.723 |
| 23 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
1.012 |
2.607 |
| 24 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
1.012 |
6.988 |
| 25 |
252345_at |
AT3G48640
|
unknown |
0.992 |
4.783 |
| 26 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.978 |
-1.827 |
| 27 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.975 |
4.838 |
| 28 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.973 |
7.267 |
| 29 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.959 |
8.498 |
| 30 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.945 |
5.845 |
| 31 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.933 |
4.809 |
| 32 |
244994_at |
ATCG01010
|
NDHF |
0.922 |
0.359 |
| 33 |
259385_at |
AT1G13470
|
unknown |
0.921 |
2.884 |
| 34 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.910 |
7.037 |
| 35 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.901 |
0.782 |
| 36 |
246419_at |
AT5G17030
|
UGT78D3, UDP-glucosyl transferase 78D3 |
0.894 |
-0.462 |
| 37 |
251832_at |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
0.886 |
6.516 |
| 38 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.880 |
6.024 |
| 39 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.879 |
0.583 |
| 40 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.879 |
-0.027 |
| 41 |
248123_at |
AT5G54720
|
[Ankyrin repeat family protein] |
0.871 |
3.737 |
| 42 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.871 |
7.024 |
| 43 |
248486_at |
AT5G51060
|
ATRBOHC, A. THALIANA RESPIRATORY BURST OXIDASE HOMOLOG C, RBOHC, RESPIRATORY BURST OXIDASE HOMOLOG C, RHD2, ROOT HAIR DEFECTIVE 2 |
0.871 |
8.463 |
| 44 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.856 |
3.289 |
| 45 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.849 |
-0.971 |
| 46 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.849 |
6.046 |
| 47 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.848 |
6.685 |
| 48 |
256177_at |
AT1G51620
|
[Protein kinase superfamily protein] |
0.843 |
4.805 |
| 49 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.833 |
7.672 |
| 50 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.824 |
0.520 |
| 51 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.823 |
5.306 |
| 52 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.821 |
7.414 |
| 53 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.817 |
6.125 |
| 54 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.815 |
8.402 |
| 55 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.813 |
-1.206 |
| 56 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.812 |
2.900 |
| 57 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.809 |
7.945 |
| 58 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.808 |
0.133 |
| 59 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.806 |
0.222 |
| 60 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
0.803 |
6.788 |
| 61 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.801 |
7.038 |
| 62 |
256114_at |
AT1G16850
|
unknown |
0.796 |
-3.307 |
| 63 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.795 |
-0.573 |
| 64 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.790 |
5.376 |
| 65 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.787 |
6.916 |
| 66 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
0.785 |
-0.248 |
| 67 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.779 |
4.529 |
| 68 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
0.778 |
6.481 |
| 69 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.776 |
7.415 |
| 70 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.775 |
6.362 |
| 71 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.772 |
5.999 |
| 72 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.772 |
-0.618 |
| 73 |
262085_at |
AT1G56060
|
unknown |
0.772 |
9.872 |
| 74 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.767 |
0.640 |
| 75 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.763 |
4.995 |
| 76 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.762 |
0.524 |
| 77 |
261836_at |
AT1G16090
|
WAKL7, wall associated kinase-like 7 |
0.760 |
4.587 |
| 78 |
258203_at |
AT3G13950
|
unknown |
0.753 |
5.327 |
| 79 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.748 |
5.985 |
| 80 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.748 |
-1.121 |
| 81 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.742 |
-0.308 |
| 82 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.740 |
7.239 |
| 83 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.737 |
5.591 |
| 84 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.737 |
-0.336 |
| 85 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.735 |
2.969 |
| 86 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.735 |
6.595 |
| 87 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.728 |
3.252 |
| 88 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.725 |
0.625 |
| 89 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.715 |
5.591 |
| 90 |
267036_at |
AT2G38465
|
unknown |
0.714 |
1.314 |
| 91 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.710 |
6.643 |
| 92 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
0.703 |
3.352 |
| 93 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.703 |
3.012 |
| 94 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
0.703 |
-0.267 |
| 95 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.696 |
0.048 |
| 96 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.695 |
0.279 |
| 97 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.694 |
5.819 |
| 98 |
259928_at |
AT1G34380
|
[5'-3' exonuclease family protein] |
0.691 |
0.722 |
| 99 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.688 |
7.728 |
| 100 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.688 |
5.521 |
| 101 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.684 |
2.422 |
| 102 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
0.679 |
4.592 |
| 103 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.675 |
0.431 |
| 104 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.674 |
-0.344 |
| 105 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
0.673 |
-0.751 |
| 106 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.673 |
4.716 |
| 107 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.672 |
7.543 |
| 108 |
263221_at |
AT1G30620
|
HSR8, HIGH SUGAR RESPONSE8, MUR4, MURUS 4, UXE1, UDP-D-XYLOSE 4-EPIMERASE 1 |
0.670 |
4.844 |
| 109 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.668 |
5.271 |
| 110 |
256924_at |
AT3G29590
|
AT5MAT |
0.668 |
-1.048 |
| 111 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
0.667 |
-0.693 |
| 112 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.665 |
4.923 |
| 113 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.660 |
4.497 |
| 114 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.660 |
-0.995 |
| 115 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.659 |
-0.325 |
| 116 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.659 |
-0.499 |
| 117 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.658 |
5.033 |
| 118 |
254060_at |
AT4G25350
|
SHB1, SHORT HYPOCOTYL UNDER BLUE1 |
0.655 |
1.982 |
| 119 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
0.653 |
0.405 |
| 120 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
0.653 |
1.949 |
| 121 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.651 |
-0.144 |
| 122 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.650 |
6.216 |
| 123 |
265931_at |
AT2G18520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.649 |
-0.179 |
| 124 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.637 |
2.306 |
| 125 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.637 |
5.041 |
| 126 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.636 |
5.511 |
| 127 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.636 |
5.301 |
| 128 |
260005_at |
AT1G67920
|
unknown |
0.634 |
5.166 |
| 129 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
0.632 |
2.736 |
| 130 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.630 |
-0.106 |
| 131 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.627 |
5.743 |
| 132 |
247064_at |
AT5G66890
|
[Leucine-rich repeat (LRR) family protein] |
0.626 |
8.191 |
| 133 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.625 |
3.928 |
| 134 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
0.624 |
4.176 |
| 135 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.623 |
-0.555 |
| 136 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.615 |
4.884 |
| 137 |
247410_at |
AT5G62990
|
emb1692, embryo defective 1692 |
0.615 |
-0.235 |
| 138 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.614 |
4.700 |
| 139 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.614 |
-0.604 |
| 140 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
0.611 |
-0.648 |
| 141 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
0.611 |
-0.048 |
| 142 |
261124_at |
AT1G04900
|
unknown |
0.610 |
-0.331 |
| 143 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
0.610 |
7.417 |
| 144 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.609 |
6.198 |
| 145 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.606 |
3.199 |
| 146 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.603 |
3.375 |
| 147 |
254080_at |
AT4G25630
|
ATFIB2, FIB2, fibrillarin 2 |
0.601 |
-0.605 |
| 148 |
251476_at |
AT3G59670
|
unknown |
0.593 |
-0.533 |
| 149 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.592 |
0.099 |
| 150 |
257487_at |
AT1G71850
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.586 |
-0.718 |
| 151 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
0.585 |
1.081 |
| 152 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.582 |
4.211 |
| 153 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.581 |
3.550 |
| 154 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.580 |
4.789 |
| 155 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.580 |
0.225 |
| 156 |
267309_at |
AT2G19385
|
[zinc ion binding] |
0.579 |
0.148 |
| 157 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
0.577 |
5.396 |
| 158 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.575 |
4.337 |
| 159 |
258254_at |
AT3G26782
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.575 |
-0.151 |
| 160 |
245401_at |
AT4G17670
|
unknown |
0.574 |
2.070 |
| 161 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.572 |
-0.125 |
| 162 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.572 |
5.748 |
| 163 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
0.571 |
-0.197 |
| 164 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.570 |
3.218 |
| 165 |
253605_at |
AT4G30990
|
[ARM repeat superfamily protein] |
0.566 |
-0.340 |
| 166 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.565 |
3.733 |
| 167 |
264948_at |
AT1G77030
|
[hydrolases, acting on acid anhydrides, in phosphorus-containing anhydrides] |
0.564 |
-0.178 |
| 168 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.561 |
-0.019 |
| 169 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
0.561 |
-0.935 |
| 170 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.560 |
-0.398 |
| 171 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.560 |
5.525 |
| 172 |
247959_at |
AT5G57080
|
unknown |
0.558 |
-0.318 |
| 173 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.557 |
6.582 |
| 174 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
0.557 |
1.878 |
| 175 |
254857_at |
AT4G12120
|
ATSEC1B, SEC1B |
0.557 |
4.361 |
| 176 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.554 |
0.619 |
| 177 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
0.553 |
4.213 |
| 178 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.551 |
-0.064 |
| 179 |
265061_at |
AT1G61640
|
[Protein kinase superfamily protein] |
0.551 |
2.920 |
| 180 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.550 |
1.923 |
| 181 |
262777_at |
AT1G13030
|
Atcoilin, COILIN, coilin |
0.549 |
-0.480 |
| 182 |
251611_at |
AT3G57940
|
unknown |
0.549 |
-0.459 |
| 183 |
258166_at |
AT3G21540
|
[transducin family protein / WD-40 repeat family protein] |
0.541 |
-0.420 |
| 184 |
248605_at |
AT5G49410
|
unknown |
0.539 |
-0.320 |
| 185 |
256858_at |
AT3G15140
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.538 |
-0.455 |
| 186 |
248510_at |
AT5G50315
|
[Mutator-like transposase family, has a 5.0e-14 P-value blast match to Q9XE24 /118-277 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.537 |
-0.323 |
| 187 |
257193_at |
AT3G13160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.536 |
-0.289 |
| 188 |
266231_at |
AT2G02220
|
ATPSKR1, PHYTOSULFOKIN RECEPTOR 1, PSKR1, phytosulfokin receptor 1 |
0.534 |
3.156 |
| 189 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.530 |
-0.285 |
| 190 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
0.529 |
-0.562 |
| 191 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.528 |
3.858 |
| 192 |
254847_at |
AT4G11850
|
MEE54, maternal effect embryo arrest 54, PLDGAMMA1, phospholipase D gamma 1 |
0.528 |
3.704 |
| 193 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
0.528 |
2.985 |
| 194 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.528 |
5.850 |
| 195 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.527 |
-0.832 |
| 196 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.527 |
2.786 |
| 197 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
0.526 |
-0.311 |
| 198 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.526 |
2.311 |
| 199 |
247367_at |
AT5G63290
|
[Radical SAM superfamily protein] |
0.525 |
-0.837 |
| 200 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.524 |
5.282 |
| 201 |
251090_at |
AT5G01340
|
AtmSFC1, mSFC1, mitochondrial succinate-fumarate carrier 1 |
0.523 |
3.311 |
| 202 |
251633_at |
AT3G57460
|
[catalytics] |
0.523 |
4.011 |
| 203 |
259721_at |
AT1G60890
|
[Phosphatidylinositol-4-phosphate 5-kinase family protein] |
0.522 |
-0.106 |
| 204 |
267004_at |
AT2G34260
|
WDR55, human WDR55 (WD40 repeat) homolog |
0.521 |
-0.322 |
| 205 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.520 |
3.857 |
| 206 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.519 |
3.210 |
| 207 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
0.517 |
0.059 |
| 208 |
256230_at |
AT3G12340
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.516 |
0.512 |
| 209 |
260688_at |
AT1G17665
|
unknown |
0.516 |
1.763 |
| 210 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.516 |
2.513 |
| 211 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.515 |
1.943 |
| 212 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.514 |
-0.512 |
| 213 |
257724_at |
AT3G18510
|
unknown |
0.514 |
-0.536 |
| 214 |
259213_at |
AT3G09010
|
[Protein kinase superfamily protein] |
0.513 |
4.798 |
| 215 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
0.511 |
0.456 |
| 216 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.511 |
2.209 |
| 217 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
0.511 |
0.018 |
| 218 |
263854_at |
AT2G04430
|
atnudt5, nudix hydrolase homolog 5, NUDT5, nudix hydrolase homolog 5 |
0.510 |
2.907 |
| 219 |
265154_at |
AT1G30960
|
[GTP-binding family protein] |
0.510 |
-0.380 |
| 220 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.509 |
4.266 |
| 221 |
259503_at |
AT1G15870
|
[Mitochondrial glycoprotein family protein] |
0.508 |
0.780 |
| 222 |
265326_at |
AT2G18220
|
[Noc2p family] |
0.507 |
0.384 |
| 223 |
250787_at |
AT5G05560
|
APC1, anaphase promoting complex 1, EMB2771, EMBRYO DEFECTIVE 2771 |
0.507 |
-0.402 |
| 224 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.507 |
-0.545 |
| 225 |
266017_at |
AT2G18690
|
unknown |
0.506 |
7.480 |
| 226 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.504 |
2.090 |
| 227 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.503 |
-0.487 |
| 228 |
258545_at |
AT3G07050
|
NSN1, nucleostemin-like 1 |
0.501 |
-0.242 |
| 229 |
245034_at |
AT2G26390
|
[Serine protease inhibitor (SERPIN) family protein] |
0.500 |
3.629 |
| 230 |
245000_at |
ATCG00210
|
YCF6 |
0.500 |
-0.701 |
| 231 |
251985_at |
AT3G53220
|
[Thioredoxin superfamily protein] |
0.499 |
-0.026 |
| 232 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.497 |
-0.153 |
| 233 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
0.497 |
-0.375 |
| 234 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.495 |
-0.757 |
| 235 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.495 |
0.242 |
| 236 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.494 |
3.639 |
| 237 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.490 |
1.096 |
| 238 |
258316_at |
AT3G22660
|
[rRNA processing protein-related] |
0.490 |
0.216 |
| 239 |
251678_at |
AT3G56990
|
EDA7, embryo sac development arrest 7 |
0.488 |
-0.243 |
| 240 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.488 |
2.998 |
| 241 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
0.488 |
1.437 |
| 242 |
256284_at |
AT3G12530
|
PSF2 |
0.486 |
-0.389 |
| 243 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
0.484 |
-0.761 |
| 244 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.483 |
-2.005 |
| 245 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.483 |
4.483 |
| 246 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.482 |
1.826 |
| 247 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.481 |
-0.364 |
| 248 |
246767_at |
AT5G27395
|
[Mitochondrial inner membrane translocase complex, subunit Tim44-related protein] |
0.480 |
-0.155 |
| 249 |
261259_at |
AT1G26660
|
[Prefoldin chaperone subunit family protein] |
0.480 |
0.211 |
| 250 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.479 |
-0.933 |
| 251 |
262611_at |
AT1G14060
|
[GCK domain-containing protein] |
0.479 |
1.919 |
| 252 |
249252_at |
AT5G42010
|
[Transducin/WD40 repeat-like superfamily protein] |
0.478 |
4.579 |
| 253 |
249004_at |
AT5G44570
|
unknown |
0.476 |
2.756 |
| 254 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.476 |
0.114 |
| 255 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.476 |
4.856 |
| 256 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.475 |
4.569 |
| 257 |
257765_at |
AT3G23020
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.475 |
-0.587 |
| 258 |
265078_at |
AT1G55500
|
ECT4, evolutionarily conserved C-terminal region 4 |
0.475 |
1.639 |
| 259 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
0.474 |
0.788 |
| 260 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
0.474 |
0.135 |
| 261 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.474 |
4.189 |
| 262 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.474 |
4.605 |
| 263 |
267474_at |
AT2G02740
|
ATWHY3, A. THALIANA WHIRLY 3, PTAC11, PLASTID TRANSCRIPTIONALLY ACTIVE11, WHY3, WHIRLY 3 |
0.473 |
-0.504 |
| 264 |
258202_at |
AT3G13940
|
[DNA binding] |
0.473 |
0.520 |
| 265 |
253490_at |
AT4G31790
|
[Tetrapyrrole (Corrin/Porphyrin) Methylases] |
0.473 |
1.149 |
| 266 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.472 |
0.222 |
| 267 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.472 |
-1.542 |
| 268 |
258220_at |
AT3G17830
|
DJA4, DNA J protein A4 |
0.472 |
-1.091 |
| 269 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.472 |
1.698 |
| 270 |
266076_at |
AT2G40700
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.469 |
-0.492 |
| 271 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.468 |
6.089 |
| 272 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.467 |
5.882 |
| 273 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.466 |
-0.667 |
| 274 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
0.466 |
-0.177 |
| 275 |
256065_at |
AT1G07070
|
[Ribosomal protein L35Ae family protein] |
0.466 |
-0.699 |
| 276 |
258009_at |
AT3G19440
|
[Pseudouridine synthase family protein] |
0.466 |
-0.071 |
| 277 |
262584_at |
AT1G15440
|
ATPWP2, PERIODIC TRYPTOPHAN PROTEIN 2, PWP2, periodic tryptophan protein 2 |
0.466 |
-0.205 |
| 278 |
263681_at |
AT1G26840
|
ATORC6, ARABIDOPSIS THALIANA ORIGIN RECOGNITION COMPLEX PROTEIN 6, ORC6, origin recognition complex protein 6 |
0.464 |
0.297 |
| 279 |
264818_at |
AT1G03530
|
ATNAF1, NAF1, nuclear assembly factor 1 |
0.463 |
-0.559 |
| 280 |
251530_at |
AT3G58520
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.462 |
-0.189 |
| 281 |
255501_at |
AT4G02400
|
[U3 ribonucleoprotein (Utp) family protein] |
0.460 |
0.380 |
| 282 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
0.460 |
0.602 |
| 283 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
0.460 |
0.001 |
| 284 |
264951_at |
AT1G76970
|
[Target of Myb protein 1] |
0.459 |
4.013 |
| 285 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
0.458 |
-0.390 |
| 286 |
254208_at |
AT4G24175
|
unknown |
0.458 |
-0.566 |
| 287 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.455 |
-0.146 |
| 288 |
249314_at |
AT5G41180
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.454 |
2.308 |
| 289 |
245518_at |
AT4G15850
|
ATRH1, RNA helicase 1, RH1, RNA helicase 1 |
0.454 |
-0.164 |
| 290 |
246052_at |
AT5G08305
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.454 |
-0.102 |
| 291 |
251005_at |
AT5G02590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.453 |
0.225 |
| 292 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
0.451 |
-0.777 |
| 293 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
0.451 |
2.167 |
| 294 |
263474_at |
AT2G31725
|
unknown |
0.450 |
0.481 |
| 295 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
0.450 |
-0.316 |
| 296 |
253981_at |
AT4G26670
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
0.449 |
-0.818 |
| 297 |
256756_at |
AT3G25610
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.449 |
5.381 |
| 298 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
0.449 |
-1.590 |
| 299 |
253137_at |
AT4G35500
|
[Protein kinase superfamily protein] |
0.449 |
1.925 |
| 300 |
248410_at |
AT5G51570
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.449 |
1.802 |
| 301 |
256096_at |
AT1G13650
|
unknown |
-1.119 |
-4.163 |
| 302 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-1.092 |
-3.410 |
| 303 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-1.088 |
-0.308 |
| 304 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-1.023 |
-3.706 |
| 305 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.923 |
-1.813 |
| 306 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.854 |
-0.627 |
| 307 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.764 |
-1.319 |
| 308 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.734 |
-0.797 |
| 309 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.733 |
-0.263 |
| 310 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.728 |
3.100 |
| 311 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.726 |
-0.453 |
| 312 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.709 |
-2.583 |
| 313 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.696 |
-2.068 |
| 314 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.693 |
-1.581 |
| 315 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.683 |
-4.674 |
| 316 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.682 |
-3.894 |
| 317 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.669 |
-1.449 |
| 318 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.669 |
-3.125 |
| 319 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.667 |
0.158 |
| 320 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
-0.662 |
0.449 |
| 321 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.662 |
-2.050 |
| 322 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.657 |
-2.792 |
| 323 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.626 |
-2.512 |
| 324 |
260877_at |
AT1G21500
|
unknown |
-0.623 |
-1.119 |
| 325 |
253305_at |
AT4G33666
|
unknown |
-0.618 |
-2.314 |
| 326 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
-0.618 |
-1.669 |
| 327 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.616 |
0.998 |
| 328 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.616 |
-3.095 |
| 329 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.609 |
2.276 |
| 330 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.608 |
-1.478 |
| 331 |
255957_at |
AT1G22160
|
unknown |
-0.607 |
-1.852 |
| 332 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.607 |
1.072 |
| 333 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
-0.602 |
-1.006 |
| 334 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.598 |
-0.598 |
| 335 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.597 |
-3.020 |
| 336 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.594 |
-0.293 |
| 337 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.592 |
-2.487 |
| 338 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.588 |
-1.837 |
| 339 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.587 |
0.225 |
| 340 |
254787_at |
AT4G12690
|
unknown |
-0.585 |
0.008 |
| 341 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.583 |
-2.579 |
| 342 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.583 |
-1.084 |
| 343 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.577 |
-0.595 |
| 344 |
264343_at |
AT1G11850
|
unknown |
-0.575 |
-0.422 |
| 345 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.572 |
-1.128 |
| 346 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.564 |
-2.584 |
| 347 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.563 |
-0.945 |
| 348 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.562 |
-2.852 |
| 349 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.562 |
-1.321 |
| 350 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.562 |
-1.456 |
| 351 |
263118_at |
AT1G03090
|
MCCA |
-0.561 |
-0.237 |
| 352 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.561 |
-1.145 |
| 353 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.559 |
-1.353 |
| 354 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.559 |
-0.924 |
| 355 |
256603_at |
AT3G28270
|
unknown |
-0.558 |
-3.138 |
| 356 |
253701_at |
AT4G29890
|
[choline monooxygenase, putative (CMO-like)] |
-0.557 |
-0.625 |
| 357 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.555 |
-1.924 |
| 358 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.552 |
-1.852 |
| 359 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.551 |
-1.214 |
| 360 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.550 |
-2.149 |
| 361 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.545 |
-2.114 |
| 362 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.544 |
0.546 |
| 363 |
249378_at |
AT5G40450
|
unknown |
-0.540 |
0.192 |
| 364 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.539 |
-2.907 |
| 365 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.538 |
0.190 |
| 366 |
264774_at |
AT1G22890
|
unknown |
-0.538 |
4.693 |
| 367 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.538 |
0.001 |
| 368 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.538 |
-2.312 |
| 369 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
-0.536 |
-1.961 |
| 370 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.535 |
1.095 |
| 371 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.534 |
-1.962 |
| 372 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.532 |
-0.219 |
| 373 |
266324_at |
AT2G46710
|
ROPGAP3, ROP guanosine triphosphatase (GTPase)-activating protein 3 |
-0.530 |
-1.612 |
| 374 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.528 |
-1.697 |
| 375 |
260317_at |
AT1G63800
|
UBC5, ubiquitin-conjugating enzyme 5 |
-0.524 |
-1.158 |
| 376 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.523 |
-0.308 |
| 377 |
267538_at |
AT2G41870
|
[Remorin family protein] |
-0.523 |
-2.147 |
| 378 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.522 |
-0.343 |
| 379 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.520 |
-1.739 |
| 380 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
-0.518 |
-0.244 |
| 381 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.518 |
-1.142 |
| 382 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.517 |
-0.543 |
| 383 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.516 |
-0.082 |
| 384 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.515 |
-3.989 |
| 385 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.513 |
-2.819 |
| 386 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.510 |
1.098 |
| 387 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
-0.507 |
-0.471 |
| 388 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.500 |
0.961 |
| 389 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.498 |
-1.206 |
| 390 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-0.495 |
0.463 |
| 391 |
247455_at |
AT5G62470
|
ATMYB96, MYB DOMAIN PROTEIN 96, MYB96, myb domain protein 96, MYBCOV1 |
-0.494 |
-0.758 |
| 392 |
259103_at |
AT3G11690
|
unknown |
-0.493 |
-0.098 |
| 393 |
248205_at |
AT5G54300
|
unknown |
-0.491 |
-0.243 |
| 394 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
-0.491 |
-0.363 |
| 395 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
-0.490 |
-0.349 |
| 396 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.486 |
0.135 |
| 397 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.481 |
-3.990 |
| 398 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.479 |
0.526 |
| 399 |
253478_at |
AT4G32350
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.473 |
-0.906 |
| 400 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.469 |
0.432 |
| 401 |
255008_at |
AT4G10060
|
[Beta-glucosidase, GBA2 type family protein] |
-0.468 |
-1.334 |
| 402 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
-0.468 |
-1.170 |
| 403 |
254688_at |
AT4G13830
|
DJC26, DNA J protein C26, J20, DNAJ-like 20 |
-0.468 |
-0.830 |
| 404 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.467 |
-1.595 |
| 405 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
-0.466 |
-1.236 |
| 406 |
250156_at |
AT5G15170
|
TDP1, tyrosyl-DNA phosphodiesterase 1 |
-0.466 |
-0.836 |
| 407 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.464 |
-0.283 |
| 408 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
-0.463 |
-1.749 |
| 409 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
-0.463 |
-1.854 |
| 410 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.461 |
-0.644 |
| 411 |
246487_at |
AT5G16030
|
unknown |
-0.459 |
-2.979 |
| 412 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.458 |
-0.132 |
| 413 |
252178_at |
AT3G50750
|
BEH1, BES1/BZR1 homolog 1 |
-0.456 |
-1.185 |
| 414 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.455 |
-2.212 |
| 415 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.452 |
-3.462 |
| 416 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.449 |
-3.188 |
| 417 |
262913_at |
AT1G59960
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.448 |
0.418 |
| 418 |
267344_at |
AT2G44230
|
unknown |
-0.447 |
-3.245 |
| 419 |
264250_at |
AT1G78680
|
ATGGH2, gamma-glutamyl hydrolase 2, GGH2, gamma-glutamyl hydrolase 2 |
-0.446 |
-1.894 |
| 420 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.446 |
-0.889 |
| 421 |
258054_at |
AT3G16240
|
AQP1, ATTIP2;1, DELTA-TIP, delta tonoplast integral protein, DELTA-TIP1, TIP2;1 |
-0.445 |
-1.314 |
| 422 |
258109_at |
AT3G23640
|
HGL1, heteroglycan glucosidase 1 |
-0.443 |
-0.985 |
| 423 |
257137_at |
AT3G28860
|
ABCB19, ATP-binding cassette B19, ATABCB19, Arabidopsis thaliana ATP-binding cassette B19, ATMDR1, ATMDR11, MULTIDRUG RESISTANCE PROTEIN 11, ATPGP19, MDR1, MDR11, PGP19, P-GLYCOPROTEIN 19 |
-0.442 |
-0.671 |
| 424 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.441 |
-0.232 |
| 425 |
255585_at |
AT4G01550
|
ANAC069, NAC domain containing protein 69, NAC069, NAC domain containing protein 69, NTM2, NAC with Transmembrane Motif 2 |
-0.441 |
0.202 |
| 426 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.436 |
-2.275 |
| 427 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.435 |
-0.419 |
| 428 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.435 |
4.520 |
| 429 |
265149_at |
AT1G51400
|
[Photosystem II 5 kD protein] |
-0.434 |
-1.974 |
| 430 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.433 |
-0.707 |
| 431 |
259431_at |
AT1G01620
|
PIP1;3, PLASMA MEMBRANE INTRINSIC PROTEIN 1;3, PIP1C, plasma membrane intrinsic protein 1C, TMP-B |
-0.433 |
-0.958 |
| 432 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.431 |
-1.282 |
| 433 |
247881_at |
AT5G57700
|
[BNR/Asp-box repeat family protein] |
-0.431 |
-0.581 |
| 434 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
-0.431 |
-1.918 |
| 435 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.430 |
-2.393 |
| 436 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.430 |
-3.475 |
| 437 |
245112_at |
AT2G41540
|
GPDHC1 |
-0.429 |
-0.598 |
| 438 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.428 |
-0.618 |
| 439 |
249355_at |
AT5G40500
|
unknown |
-0.427 |
-0.482 |
| 440 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.426 |
-1.353 |
| 441 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.425 |
-0.684 |
| 442 |
247374_at |
AT5G63190
|
[MA3 domain-containing protein] |
-0.425 |
-0.054 |
| 443 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-0.424 |
-1.180 |
| 444 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.424 |
-1.606 |
| 445 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
-0.424 |
-0.698 |
| 446 |
250569_at |
AT5G08130
|
BIM1 |
-0.423 |
-0.434 |
| 447 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.423 |
0.484 |
| 448 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
-0.422 |
0.527 |
| 449 |
261056_at |
AT1G01360
|
PYL9, PYRABACTIN RESISTANCE 1-LIKE 9, RCAR1, regulatory component of ABA receptor 1 |
-0.421 |
-0.839 |
| 450 |
263867_at |
AT2G36830
|
GAMMA-TIP, gamma tonoplast intrinsic protein, GAMMA-TIP1, GAMMA TONOPLAST INTRINSIC PROTEIN 1, TIP1;1, TONOPLAST INTRINSIC PROTEIN 1;1 |
-0.419 |
-1.435 |
| 451 |
265411_at |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.415 |
-0.273 |
| 452 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.414 |
-2.564 |
| 453 |
257875_at |
AT3G17120
|
unknown |
-0.414 |
-1.509 |
| 454 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
-0.412 |
0.043 |
| 455 |
265913_at |
AT2G25625
|
unknown |
-0.412 |
-1.699 |
| 456 |
251142_at |
AT5G01015
|
unknown |
-0.411 |
-2.237 |
| 457 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.411 |
-0.439 |
| 458 |
265277_at |
AT2G28410
|
unknown |
-0.408 |
-2.124 |
| 459 |
255824_at |
AT2G40530
|
unknown |
-0.408 |
-0.782 |
| 460 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.407 |
-0.570 |
| 461 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
-0.405 |
2.434 |
| 462 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.403 |
-1.479 |
| 463 |
253243_at |
AT4G34560
|
unknown |
-0.403 |
-1.128 |
| 464 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-0.401 |
-0.297 |
| 465 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.401 |
-1.149 |
| 466 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
-0.399 |
-2.829 |
| 467 |
252829_at |
AT4G40060
|
ATHB-16, ATHB16, homeobox protein 16, HB16, homeobox protein 16 |
-0.397 |
-1.435 |
| 468 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.396 |
-1.598 |
| 469 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.394 |
-1.374 |
| 470 |
246097_at |
AT5G20270
|
HHP1, heptahelical transmembrane protein1 |
-0.393 |
-2.532 |
| 471 |
257789_at |
AT3G27020
|
AtYSL6, YSL6, YELLOW STRIPE like 6 |
-0.393 |
-1.242 |
| 472 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.392 |
-0.381 |
| 473 |
259366_at |
AT1G13280
|
AOC4, allene oxide cyclase 4 |
-0.392 |
-0.934 |
| 474 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.392 |
-0.296 |
| 475 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
-0.390 |
-1.752 |
| 476 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.390 |
0.184 |
| 477 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.390 |
4.716 |
| 478 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-0.389 |
-1.427 |
| 479 |
248969_at |
AT5G45310
|
unknown |
-0.389 |
-1.902 |
| 480 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
-0.389 |
0.057 |
| 481 |
264517_at |
AT1G10120
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.388 |
-0.294 |
| 482 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
-0.387 |
-2.109 |
| 483 |
264738_at |
AT1G62250
|
unknown |
-0.387 |
-1.413 |
| 484 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.386 |
-1.870 |
| 485 |
247356_at |
AT5G63800
|
BGAL6, beta-galactosidase 6, MUM2, MUCILAGE-MODIFIED 2 |
-0.386 |
-1.180 |
| 486 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.385 |
-1.715 |
| 487 |
259012_at |
AT3G07360
|
ATPUB9, ARABIDOPSIS THALIANA PLANT U-BOX 9, PUB9, plant U-box 9 |
-0.385 |
-1.934 |
| 488 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.384 |
-0.455 |
| 489 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.382 |
-2.921 |
| 490 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
-0.382 |
-1.562 |
| 491 |
252866_at |
AT4G39840
|
unknown |
-0.382 |
-0.333 |
| 492 |
262411_at |
AT1G34640
|
[peptidases] |
-0.382 |
-1.636 |
| 493 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.381 |
-5.556 |
| 494 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
-0.377 |
-1.473 |
| 495 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
-0.376 |
0.835 |
| 496 |
261151_at |
AT1G19650
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.376 |
-0.128 |
| 497 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.376 |
-1.251 |
| 498 |
265118_at |
AT1G62660
|
[Glycosyl hydrolases family 32 protein] |
-0.375 |
-1.204 |
| 499 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.374 |
0.251 |
| 500 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
-0.372 |
-1.750 |
| 501 |
259163_at |
AT3G01490
|
[Protein kinase superfamily protein] |
-0.371 |
-0.589 |
| 502 |
259984_at |
AT1G76460
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.370 |
-0.743 |
| 503 |
250017_at |
AT5G18140
|
DJC69, DNA J protein A69 |
-0.370 |
-1.195 |
| 504 |
262049_at |
AT1G80180
|
unknown |
-0.370 |
0.555 |
| 505 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.369 |
-1.611 |
| 506 |
261559_at |
AT1G01780
|
PLIM2b, PLIM2b |
-0.369 |
-0.619 |
| 507 |
247396_at |
AT5G62930
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.368 |
-0.310 |
| 508 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-0.368 |
0.009 |
| 509 |
253886_at |
AT4G27710
|
CYP709B3, cytochrome P450, family 709, subfamily B, polypeptide 3 |
-0.368 |
-1.858 |
| 510 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.367 |
-2.738 |
| 511 |
251493_at |
AT3G59300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.366 |
-1.198 |
| 512 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
-0.366 |
-1.003 |
| 513 |
252646_at |
AT3G44610
|
[Protein kinase superfamily protein] |
-0.366 |
-2.150 |
| 514 |
253877_at |
AT4G27435
|
unknown |
-0.365 |
-0.202 |
| 515 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
-0.363 |
-0.345 |
| 516 |
248509_at |
AT5G50335
|
unknown |
-0.361 |
1.427 |
| 517 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.360 |
0.965 |
| 518 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.358 |
-2.166 |
| 519 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.357 |
1.004 |
| 520 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.357 |
2.102 |
| 521 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
-0.356 |
-0.833 |
| 522 |
263827_at |
AT2G40420
|
[Transmembrane amino acid transporter family protein] |
-0.356 |
-1.072 |
| 523 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
-0.356 |
-1.371 |
| 524 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.355 |
-1.312 |
| 525 |
250556_at |
AT5G07920
|
ATDGK1, DIACYLGLYCEROL KINASE 1, DGK1, diacylglycerol kinase1 |
-0.354 |
-0.347 |
| 526 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.354 |
-0.286 |
| 527 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
-0.353 |
-0.483 |
| 528 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
-0.353 |
-1.515 |
| 529 |
250016_at |
AT5G18100
|
CSD3, copper/zinc superoxide dismutase 3 |
-0.353 |
-0.938 |
| 530 |
250470_at |
AT5G10160
|
[Thioesterase superfamily protein] |
-0.353 |
-0.069 |
| 531 |
254362_at |
AT4G22160
|
unknown |
-0.352 |
-0.165 |
| 532 |
263035_at |
AT1G23860
|
At-RSZ21, RS-containing zinc finger protein 21, RSZ21, RS-containing zinc finger protein 21, RSZP21, RS-containing zinc finger protein 21, SRZ-21, SRZ21 |
-0.351 |
0.144 |
| 533 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.350 |
0.850 |
| 534 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.349 |
-2.551 |
| 535 |
257894_at |
AT3G17100
|
AIF3, ATBS1 Interacting Factor 3 |
-0.349 |
-0.019 |
| 536 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.349 |
0.068 |
| 537 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
-0.349 |
0.081 |
| 538 |
246909_at |
AT5G25770
|
[alpha/beta-Hydrolases superfamily protein] |
-0.348 |
1.373 |
| 539 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.348 |
-1.462 |
| 540 |
249122_at |
AT5G43850
|
ARD4, ATARD4 |
-0.347 |
-2.003 |
| 541 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.346 |
-1.769 |
| 542 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.346 |
-1.919 |
| 543 |
249693_at |
AT5G35750
|
AHK2, histidine kinase 2, HK2, histidine kinase 2 |
-0.346 |
-1.286 |
| 544 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.344 |
-1.676 |
| 545 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.344 |
-2.288 |
| 546 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
-0.343 |
-0.702 |
| 547 |
258375_at |
AT3G17470
|
ATCRSH, CRSH, Ca2+-activated RelA/spot homolog |
-0.343 |
-0.910 |
| 548 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.343 |
-0.684 |
| 549 |
258038_at |
AT3G21260
|
GLTP3, GLYCOLIPID TRANSFER PROTEIN 3 |
-0.343 |
-1.700 |
| 550 |
256674_at |
AT3G52360
|
unknown |
-0.343 |
-0.727 |
| 551 |
248167_at |
AT5G54530
|
unknown |
-0.342 |
-0.942 |
| 552 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.342 |
-3.560 |
| 553 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.341 |
-0.694 |
| 554 |
249818_at |
AT5G23860
|
TUB8, tubulin beta 8 |
-0.341 |
0.684 |
| 555 |
260536_at |
AT2G43400
|
ETFQO, electron-transfer flavoprotein:ubiquinone oxidoreductase |
-0.340 |
-0.307 |
| 556 |
262558_at |
AT1G31335
|
unknown |
-0.340 |
0.035 |
| 557 |
245307_at |
AT4G16770
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.339 |
-1.527 |
| 558 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.339 |
-0.234 |
| 559 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.339 |
-2.300 |
| 560 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
-0.338 |
-0.243 |
| 561 |
264862_at |
AT1G24330
|
[ARM repeat superfamily protein] |
-0.337 |
-1.361 |
| 562 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.337 |
-0.349 |
| 563 |
258774_at |
AT3G10740
|
ARAF, ALPHA-L-ARABINOFURANOSIDASE, ARAF1, ALPHA-L-ARABINOFURANOSIDASE 1, ASD1, alpha-L-arabinofuranosidase 1, ATASD1, ARABIDOPSIS THALIANA ALPHA-L-ARABINOFURANOSIDASE 1 |
-0.337 |
-0.119 |
| 564 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.336 |
-2.189 |
| 565 |
261667_at |
AT1G18460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.335 |
-1.368 |
| 566 |
246041_at |
AT5G19290
|
[alpha/beta-Hydrolases superfamily protein] |
-0.335 |
0.192 |
| 567 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.335 |
-1.164 |
| 568 |
247072_at |
AT5G66490
|
unknown |
-0.335 |
-0.648 |
| 569 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-0.334 |
-0.836 |
| 570 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-0.334 |
-1.293 |
| 571 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.334 |
-1.762 |
| 572 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.334 |
-1.943 |
| 573 |
259605_at |
AT1G27910
|
ATPUB45, ARABIDOPSIS THALIANA PLANT U-BOX 45, PUB45, plant U-box 45 |
-0.334 |
-0.270 |
| 574 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
-0.334 |
0.048 |
| 575 |
245776_at |
AT1G30260
|
unknown |
-0.334 |
-1.398 |
| 576 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.333 |
-2.430 |
| 577 |
246783_at |
AT5G27360
|
SFP2 |
-0.333 |
-1.767 |
| 578 |
251011_at |
AT5G02560
|
HTA12, histone H2A 12 |
-0.333 |
-0.506 |
| 579 |
266188_at |
AT2G39000
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.332 |
-1.424 |
| 580 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.332 |
-2.475 |
| 581 |
254239_at |
AT4G23400
|
PIP1;5, plasma membrane intrinsic protein 1;5, PIP1D |
-0.331 |
-1.462 |
| 582 |
245078_at |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
-0.331 |
-1.477 |
| 583 |
253672_at |
AT4G29820
|
ATCFIM-25, ARABIDOPSIS THALIANA HOMOLOG OF CFIM-25, CFIM-25, homolog of CFIM-25 |
-0.330 |
0.631 |
| 584 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.330 |
-1.002 |
| 585 |
256226_at |
AT1G56280
|
ATDI19, drought-induced 19, DI19, drought-induced 19 |
-0.330 |
-0.055 |
| 586 |
258188_at |
AT3G17800
|
unknown |
-0.330 |
0.632 |
| 587 |
247031_at |
AT5G67230
|
I14H, IRREGULAR XYLEM 14 Homolog, IRX14-L, IRREGULAR XYLEM 14-LIKE |
-0.329 |
0.001 |
| 588 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
-0.329 |
-0.254 |
| 589 |
256458_at |
AT1G75220
|
AtERDL6, ERDL6, ERD6-like 6 |
-0.328 |
0.194 |
| 590 |
254608_at |
AT4G18910
|
ATNLM2, NOD26-LIKE INTRINSIC PROTEIN 2, NIP1;2, NOD26-like intrinsic protein 1;2, NLM2, NOD26-LIKE INTRINSIC PROTEIN 2 |
-0.328 |
-0.359 |
| 591 |
255602_at |
AT4G01026
|
PYL7, PYR1-like 7, RCAR2, regulatory components of ABA receptor 2 |
-0.328 |
-0.613 |
| 592 |
259674_at |
AT1G77700
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.326 |
0.268 |
| 593 |
258993_at |
AT3G08940
|
LHCB4.2, light harvesting complex photosystem II |
-0.325 |
-1.654 |
| 594 |
251391_at |
AT3G60910
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.325 |
-0.946 |
| 595 |
250694_at |
AT5G06710
|
HAT14, homeobox from Arabidopsis thaliana |
-0.325 |
-0.038 |
| 596 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
-0.323 |
-0.705 |
| 597 |
261900_at |
AT1G80940
|
unknown |
-0.323 |
-0.259 |
| 598 |
249332_at |
AT5G40980
|
unknown |
-0.323 |
0.070 |
| 599 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.322 |
-1.342 |
| 600 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.322 |
-1.192 |