|
probeID |
AGICode |
Annotation |
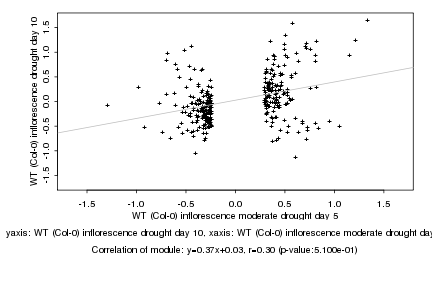
Log2 signal ratio
WT (Col-0) inflorescence moderate drought day 5 |
Log2 signal ratio
WT (Col-0) inflorescence drought day 10 |
| 1 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
1.334 |
1.664 |
| 2 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.210 |
1.241 |
| 3 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
1.147 |
0.941 |
| 4 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
1.048 |
-0.493 |
| 5 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.950 |
-0.392 |
| 6 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.837 |
-0.531 |
| 7 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.814 |
0.301 |
| 8 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.810 |
1.227 |
| 9 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.808 |
-0.433 |
| 10 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.802 |
0.944 |
| 11 |
256521_at |
AT1G66120
|
AAE11, acyl-activating enzyme 11, AtAAE11 |
0.799 |
0.823 |
| 12 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.757 |
1.069 |
| 13 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.757 |
0.268 |
| 14 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.721 |
-0.511 |
| 15 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
0.719 |
-0.588 |
| 16 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.718 |
1.082 |
| 17 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
0.712 |
-0.761 |
| 18 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.710 |
1.182 |
| 19 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.700 |
1.123 |
| 20 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.677 |
-0.405 |
| 21 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.675 |
-0.430 |
| 22 |
267476_at |
AT2G02720
|
[Pectate lyase family protein] |
0.637 |
0.815 |
| 23 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.635 |
-0.612 |
| 24 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.613 |
0.990 |
| 25 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.605 |
-0.341 |
| 26 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.602 |
-1.119 |
| 27 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
0.601 |
0.579 |
| 28 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
0.568 |
0.057 |
| 29 |
247061_at |
AT5G66780
|
unknown |
0.567 |
1.600 |
| 30 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.559 |
0.544 |
| 31 |
249134_at |
AT5G43150
|
unknown |
0.557 |
0.490 |
| 32 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.556 |
0.040 |
| 33 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
0.540 |
0.074 |
| 34 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
0.527 |
-0.505 |
| 35 |
254829_at |
AT4G12530
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.526 |
0.251 |
| 36 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.525 |
0.140 |
| 37 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.524 |
-0.036 |
| 38 |
252661_at |
AT3G44450
|
unknown |
0.523 |
-0.609 |
| 39 |
257850_at |
AT3G13065
|
SRF4, STRUBBELIG-receptor family 4 |
0.522 |
0.201 |
| 40 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
0.522 |
-0.027 |
| 41 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.517 |
0.909 |
| 42 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.511 |
-0.060 |
| 43 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
0.505 |
-0.059 |
| 44 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.501 |
0.952 |
| 45 |
256114_at |
AT1G16850
|
unknown |
0.500 |
1.342 |
| 46 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.495 |
0.169 |
| 47 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.491 |
1.175 |
| 48 |
257186_at |
AT3G13130
|
unknown |
0.487 |
1.074 |
| 49 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.487 |
-0.365 |
| 50 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.486 |
0.736 |
| 51 |
267511_at |
AT2G45670
|
[calcineurin B subunit-related] |
0.485 |
0.162 |
| 52 |
250204_at |
AT5G13990
|
ATEXO70C2, exocyst subunit exo70 family protein C2, EXO70C2, exocyst subunit exo70 family protein C2 |
0.479 |
0.214 |
| 53 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.471 |
0.551 |
| 54 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
0.460 |
0.336 |
| 55 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.460 |
0.392 |
| 56 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.455 |
0.581 |
| 57 |
261923_at |
AT1G22380
|
AtUGT85A3, UDP-glucosyl transferase 85A3, UGT85A3, UDP-glucosyl transferase 85A3 |
0.453 |
0.546 |
| 58 |
266453_at |
AT2G43230
|
[Protein kinase superfamily protein] |
0.451 |
0.314 |
| 59 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.449 |
-0.576 |
| 60 |
262378_at |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
0.446 |
0.259 |
| 61 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
0.445 |
-0.105 |
| 62 |
257349_at |
AT2G30630
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.441 |
-0.078 |
| 63 |
249935_at |
AT5G22420
|
FAR7, fatty acid reductase 7 |
0.439 |
0.083 |
| 64 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.435 |
0.241 |
| 65 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
0.435 |
0.712 |
| 66 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.435 |
-0.732 |
| 67 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.431 |
-0.116 |
| 68 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.426 |
-0.002 |
| 69 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.423 |
-0.074 |
| 70 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
0.422 |
0.330 |
| 71 |
246566_at |
AT5G14940
|
[Major facilitator superfamily protein] |
0.413 |
0.060 |
| 72 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.412 |
-0.310 |
| 73 |
260032_at |
AT1G68750
|
ATPPC4, phosphoenolpyruvate carboxylase 4, PPC4, phosphoenolpyruvate carboxylase 4 |
0.411 |
-0.018 |
| 74 |
263905_at |
AT2G36190
|
AtCWIN4, AtcwINV4, cell wall invertase 4, cwINV4, cell wall invertase 4 |
0.409 |
0.040 |
| 75 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.407 |
-0.788 |
| 76 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.406 |
0.318 |
| 77 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.406 |
0.083 |
| 78 |
249468_at |
AT5G39650
|
unknown |
0.405 |
0.165 |
| 79 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.401 |
-0.312 |
| 80 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
0.399 |
-0.036 |
| 81 |
267019_at |
AT2G39130
|
[Transmembrane amino acid transporter family protein] |
0.398 |
0.160 |
| 82 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
0.397 |
0.334 |
| 83 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
0.395 |
0.227 |
| 84 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
0.392 |
0.932 |
| 85 |
262463_at |
AT1G50310
|
ATSTP9, SUGAR TRANSPORTER 9, STP9, sugar transporter 9 |
0.392 |
-0.010 |
| 86 |
267429_at |
AT2G34850
|
MEE25, maternal effect embryo arrest 25 |
0.391 |
0.862 |
| 87 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.389 |
-0.113 |
| 88 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
0.388 |
0.549 |
| 89 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.388 |
0.932 |
| 90 |
247025_at |
AT5G67030
|
ABA1, ABA DEFICIENT 1, ATABA1, ARABIDOPSIS THALIANA ABA DEFICIENT 1, ATZEP, ARABIDOPSIS THALIANA ZEAXANTHIN EPOXIDASE, IBS3, IMPAIRED IN BABA-INDUCED STERILITY 3, LOS6, LOW EXPRESSION OF OSMOTIC STRESS-RESPONSIVE GENES 6, NPQ2, NON-PHOTOCHEMICAL QUENCHING 2, ZEP, ZEAXANTHIN EPOXIDASE |
0.385 |
-0.116 |
| 91 |
246896_at |
AT5G25550
|
[Leucine-rich repeat (LRR) family protein] |
0.384 |
0.064 |
| 92 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.381 |
0.761 |
| 93 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
0.380 |
-0.197 |
| 94 |
252073_at |
AT3G51750
|
unknown |
0.379 |
0.954 |
| 95 |
257672_at |
AT3G20300
|
unknown |
0.375 |
0.515 |
| 96 |
261863_at |
AT1G50630
|
unknown |
0.371 |
0.634 |
| 97 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
0.370 |
-0.415 |
| 98 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.369 |
-0.000 |
| 99 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
0.369 |
-0.809 |
| 100 |
262626_at |
AT1G06430
|
FTSH8, FTSH protease 8 |
0.367 |
0.242 |
| 101 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.367 |
0.377 |
| 102 |
257543_at |
AT3G28960
|
[Transmembrane amino acid transporter family protein] |
0.366 |
-0.342 |
| 103 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.365 |
0.294 |
| 104 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
0.359 |
0.211 |
| 105 |
248642_at |
AT5G49120
|
unknown |
0.358 |
0.656 |
| 106 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.358 |
-0.312 |
| 107 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.355 |
0.452 |
| 108 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.355 |
-0.503 |
| 109 |
262803_at |
AT1G21000
|
[PLATZ transcription factor family protein] |
0.351 |
0.574 |
| 110 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.350 |
1.229 |
| 111 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
0.345 |
-0.013 |
| 112 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
0.343 |
0.131 |
| 113 |
246242_at |
AT4G36600
|
[Late embryogenesis abundant (LEA) protein] |
0.343 |
0.234 |
| 114 |
254683_at |
AT4G13800
|
unknown |
0.342 |
0.267 |
| 115 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.342 |
0.283 |
| 116 |
263011_at |
AT1G23250
|
[Caleosin-related family protein] |
0.341 |
-0.096 |
| 117 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.339 |
0.387 |
| 118 |
251591_at |
AT3G57680
|
[Peptidase S41 family protein] |
0.339 |
0.083 |
| 119 |
260997_at |
AT1G26610
|
[C2H2-like zinc finger protein] |
0.339 |
-0.088 |
| 120 |
264888_at |
AT1G23070
|
unknown |
0.339 |
0.352 |
| 121 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
0.336 |
0.373 |
| 122 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.336 |
0.249 |
| 123 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
0.334 |
0.119 |
| 124 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.334 |
0.227 |
| 125 |
250903_at |
AT5G03600
|
[SGNH hydrolase-type esterase superfamily protein] |
0.332 |
0.128 |
| 126 |
263544_at |
AT2G21590
|
APL4 |
0.331 |
0.327 |
| 127 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
0.329 |
0.307 |
| 128 |
246940_at |
AT5G25400
|
[Nucleotide-sugar transporter family protein] |
0.329 |
-0.081 |
| 129 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
0.326 |
0.369 |
| 130 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.323 |
-0.051 |
| 131 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
0.323 |
0.110 |
| 132 |
247408_at |
AT5G62760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.323 |
0.087 |
| 133 |
253034_at |
AT4G38230
|
ATCPK26, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 26, CPK26, calcium-dependent protein kinase 26 |
0.321 |
0.284 |
| 134 |
258878_at |
AT3G03170
|
unknown |
0.320 |
0.867 |
| 135 |
259338_at |
AT3G03800
|
ATSYP131, SYP131, syntaxin of plants 131 |
0.317 |
0.384 |
| 136 |
257057_at |
AT3G15310
|
unknown |
0.317 |
-0.203 |
| 137 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
0.311 |
-0.088 |
| 138 |
262436_at |
AT1G47610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.310 |
0.600 |
| 139 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.310 |
-0.072 |
| 140 |
251132_at |
AT5G01200
|
[Duplicated homeodomain-like superfamily protein] |
0.308 |
0.619 |
| 141 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.307 |
0.652 |
| 142 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.307 |
-0.082 |
| 143 |
247055_at |
AT5G66740
|
unknown |
0.307 |
0.010 |
| 144 |
254607_at |
AT4G18920
|
unknown |
0.306 |
0.014 |
| 145 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
0.305 |
-0.260 |
| 146 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.305 |
-0.223 |
| 147 |
248750_at |
AT5G47530
|
[Auxin-responsive family protein] |
0.305 |
-0.126 |
| 148 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
0.304 |
-0.402 |
| 149 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
0.303 |
0.484 |
| 150 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.303 |
0.391 |
| 151 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.302 |
0.585 |
| 152 |
258451_at |
AT3G22360
|
AOX1B, alternative oxidase 1B |
0.301 |
0.074 |
| 153 |
261106_at |
AT1G62990
|
IXR11, KNAT7, KNOTTED-like homeobox of Arabidopsis thaliana 7 |
0.300 |
-0.043 |
| 154 |
252440_at |
AT3G47440
|
TIP5;1, tonoplast intrinsic protein 5;1 |
0.298 |
0.191 |
| 155 |
245290_at |
AT4G16490
|
[ARM repeat superfamily protein] |
0.296 |
0.027 |
| 156 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
0.296 |
0.026 |
| 157 |
250400_at |
AT5G10740
|
[Protein phosphatase 2C family protein] |
0.294 |
0.154 |
| 158 |
257309_at |
AT3G28150
|
AXY4L, ALTERED XYLOGLUCAN 4-LIKE, TBL22, TRICHOME BIREFRINGENCE-LIKE 22 |
0.290 |
0.303 |
| 159 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.289 |
0.226 |
| 160 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.288 |
0.033 |
| 161 |
254972_at |
AT4G10440
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.288 |
-0.005 |
| 162 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-1.297 |
-0.075 |
| 163 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.981 |
0.290 |
| 164 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.924 |
-0.519 |
| 165 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.769 |
-0.032 |
| 166 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.739 |
-0.620 |
| 167 |
258289_at |
AT3G23450
|
unknown |
-0.704 |
0.150 |
| 168 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.703 |
0.846 |
| 169 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.688 |
0.982 |
| 170 |
261979_at |
AT1G37130
|
ATNR2, ARABIDOPSIS NITRATE REDUCTASE 2, B29, CHL3, CHLORATE RESISTANT 3, NIA2, nitrate reductase 2, NIA2-1, NR, NITRATE REDUCTASE, NR2, NITRATE REDUCTASE 2 |
-0.659 |
-0.748 |
| 171 |
261946_at |
AT1G64560
|
|
-0.619 |
0.164 |
| 172 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.611 |
0.770 |
| 173 |
244997_at |
ATCG00170
|
RPOC2 |
-0.608 |
-0.064 |
| 174 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.589 |
0.656 |
| 175 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
-0.580 |
-0.517 |
| 176 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.574 |
0.500 |
| 177 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.553 |
-0.436 |
| 178 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
-0.541 |
-0.206 |
| 179 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.535 |
-0.631 |
| 180 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
-0.526 |
-0.117 |
| 181 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.515 |
1.050 |
| 182 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.513 |
-0.203 |
| 183 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.503 |
0.303 |
| 184 |
267545_at |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
-0.488 |
-0.085 |
| 185 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.485 |
-0.580 |
| 186 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.481 |
-0.266 |
| 187 |
260688_at |
AT1G17665
|
unknown |
-0.465 |
0.714 |
| 188 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
-0.462 |
-0.295 |
| 189 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.462 |
0.467 |
| 190 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.459 |
-0.235 |
| 191 |
248862_at |
AT5G46730
|
[glycine-rich protein] |
-0.456 |
0.111 |
| 192 |
253322_at |
AT4G33980
|
unknown |
-0.452 |
1.127 |
| 193 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.452 |
-0.616 |
| 194 |
244938_at |
ATCG01120
|
RPS15, chloroplast ribosomal protein S15 |
-0.449 |
-0.035 |
| 195 |
244998_at |
ATCG00180
|
RPOC1 |
-0.444 |
-0.023 |
| 196 |
261840_at |
AT1G16070
|
AtTLP8, tubby like protein 8, TLP8, tubby like protein 8 |
-0.437 |
-0.421 |
| 197 |
266358_at |
AT2G32280
|
VCC, VASCULATURE COMPLEXITY AND CONNECTIVITY |
-0.436 |
-0.039 |
| 198 |
248307_at |
AT5G52850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.431 |
-0.173 |
| 199 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
-0.427 |
-0.138 |
| 200 |
253317_at |
AT4G33960
|
unknown |
-0.424 |
0.095 |
| 201 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.424 |
-0.597 |
| 202 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.424 |
-0.697 |
| 203 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
-0.421 |
0.653 |
| 204 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.411 |
-1.046 |
| 205 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.410 |
-0.034 |
| 206 |
255446_at |
AT4G02750
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.403 |
-0.059 |
| 207 |
245461_at |
AT4G17000
|
unknown |
-0.398 |
-0.407 |
| 208 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.397 |
-0.280 |
| 209 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.393 |
-0.319 |
| 210 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
-0.393 |
0.041 |
| 211 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.390 |
-0.162 |
| 212 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.389 |
-0.573 |
| 213 |
250481_at |
AT5G10310
|
unknown |
-0.386 |
-0.369 |
| 214 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.378 |
0.321 |
| 215 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.378 |
-0.190 |
| 216 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.377 |
-0.288 |
| 217 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.377 |
0.348 |
| 218 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.377 |
-0.056 |
| 219 |
267636_at |
AT2G42110
|
unknown |
-0.370 |
-0.513 |
| 220 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.370 |
0.049 |
| 221 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
-0.359 |
0.018 |
| 222 |
251610_at |
AT3G57930
|
unknown |
-0.354 |
-0.027 |
| 223 |
247948_at |
AT5G57130
|
SMXL5, SMAX1-like 5 |
-0.354 |
-0.193 |
| 224 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
-0.352 |
0.638 |
| 225 |
260472_at |
AT1G10990
|
unknown |
-0.351 |
-0.130 |
| 226 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
-0.349 |
0.187 |
| 227 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
-0.342 |
0.238 |
| 228 |
244921_s_at |
ATMG01000
|
ORF114 |
-0.342 |
-0.163 |
| 229 |
257724_at |
AT3G18510
|
unknown |
-0.337 |
-0.095 |
| 230 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.336 |
-0.329 |
| 231 |
246525_at |
AT5G15840
|
BBX1, B-box domain protein 1, CO, CONSTANS, FG |
-0.336 |
0.655 |
| 232 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
-0.335 |
-0.147 |
| 233 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
-0.333 |
-0.133 |
| 234 |
247567_at |
AT5G61190
|
[putative endonuclease or glycosyl hydrolase with C2H2-type zinc finger domain] |
-0.332 |
-0.183 |
| 235 |
259363_at |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
-0.332 |
-0.009 |
| 236 |
265075_at |
AT1G55450
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.331 |
-0.304 |
| 237 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.330 |
-0.486 |
| 238 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.329 |
-0.302 |
| 239 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.328 |
-0.528 |
| 240 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.326 |
-0.275 |
| 241 |
262286_at |
AT1G68585
|
unknown |
-0.326 |
-0.218 |
| 242 |
258108_at |
AT3G23570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.324 |
0.116 |
| 243 |
255460_at |
AT4G02800
|
unknown |
-0.323 |
-0.635 |
| 244 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.320 |
-0.258 |
| 245 |
248329_at |
AT5G52780
|
unknown |
-0.319 |
-0.307 |
| 246 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.318 |
-0.789 |
| 247 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
-0.317 |
-0.467 |
| 248 |
251985_at |
AT3G53220
|
[Thioredoxin superfamily protein] |
-0.315 |
-0.105 |
| 249 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.312 |
-0.528 |
| 250 |
245607_at |
AT4G14330
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.311 |
-0.383 |
| 251 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
-0.309 |
-0.294 |
| 252 |
266123_at |
AT2G45180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.308 |
-0.747 |
| 253 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
-0.307 |
-0.175 |
| 254 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.306 |
-0.448 |
| 255 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
-0.302 |
-0.233 |
| 256 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.300 |
-0.303 |
| 257 |
244929_at |
ATMG00580
|
NAD4, NADH dehydrogenase subunit 4 |
-0.299 |
0.126 |
| 258 |
257619_at |
AT3G24810
|
ICK3, KRP5, kip-related protein 5 |
-0.298 |
-0.101 |
| 259 |
260905_at |
AT1G02710
|
[glycine-rich protein] |
-0.298 |
0.172 |
| 260 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.295 |
-0.648 |
| 261 |
260332_at |
AT1G70470
|
unknown |
-0.294 |
-0.165 |
| 262 |
248055_at |
AT5G55830
|
LecRK-S.7, L-type lectin receptor kinase S.7 |
-0.293 |
-0.342 |
| 263 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
-0.293 |
-0.032 |
| 264 |
261058_at |
AT1G01370
|
CENH3, CENTROMERIC HISTONE H3, HTR12 |
-0.293 |
-0.383 |
| 265 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.287 |
-0.237 |
| 266 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.285 |
-0.013 |
| 267 |
263775_at |
AT2G46410
|
CPC, CAPRICE |
-0.285 |
0.038 |
| 268 |
262558_at |
AT1G31335
|
unknown |
-0.285 |
-0.341 |
| 269 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.285 |
0.217 |
| 270 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.284 |
-0.406 |
| 271 |
260359_at |
AT1G69210
|
[Uncharacterised protein family UPF0090] |
-0.284 |
0.172 |
| 272 |
244903_at |
ATMG00660
|
ORF149 |
-0.284 |
-0.042 |
| 273 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.282 |
0.313 |
| 274 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
-0.280 |
-0.516 |
| 275 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.280 |
0.011 |
| 276 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.279 |
-0.500 |
| 277 |
251244_at |
AT3G62060
|
[Pectinacetylesterase family protein] |
-0.279 |
-0.353 |
| 278 |
249357_at |
AT5G40490
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.277 |
0.086 |
| 279 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.276 |
-0.343 |
| 280 |
248280_at |
AT5G52950
|
unknown |
-0.276 |
-0.221 |
| 281 |
262899_at |
AT1G59870
|
ABCG36, ATP-binding cassette G36, ATABCG36, Arabidopsis thaliana ATP-binding cassette G36, ATPDR8, ARABIDOPSIS PLEIOTROPIC DRUG RESISTANCE 8, PDR8, PLEIOTROPIC DRUG RESISTANCE 8, PEN3, PENETRATION 3 |
-0.275 |
0.156 |
| 282 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.274 |
-0.058 |
| 283 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.274 |
-0.232 |
| 284 |
260461_at |
AT1G10980
|
[Lung seven transmembrane receptor family protein] |
-0.272 |
-0.433 |
| 285 |
253978_at |
AT4G26660
|
unknown |
-0.271 |
-0.403 |
| 286 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.270 |
0.071 |
| 287 |
263219_at |
AT1G30600
|
[Subtilase family protein] |
-0.269 |
-0.179 |
| 288 |
263236_at |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
-0.268 |
-0.218 |
| 289 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
-0.266 |
-0.011 |
| 290 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.265 |
-0.428 |
| 291 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.265 |
-0.187 |
| 292 |
261100_at |
AT1G63020
|
NRPD1, NRPD1A, nuclear RNA polymerase D1A, POL IVA, NUCLEAR RNA POLYMERASE D 1A, SDE4, SMD2, SILENCING MOVEMENT DEFICIENT 2 |
-0.264 |
-0.120 |
| 293 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-0.263 |
-0.144 |
| 294 |
250386_at |
AT5G11510
|
AtMYB3R4, myb domain protein 3R4, MYB3R-4, myb domain protein 3r-4 |
-0.263 |
-0.374 |
| 295 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
-0.263 |
-0.514 |
| 296 |
254245_at |
AT4G23240
|
CRK16, cysteine-rich RLK (RECEPTOR-like protein kinase) 16 |
-0.262 |
-0.070 |
| 297 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
-0.261 |
-0.111 |
| 298 |
248691_at |
AT5G48310
|
unknown |
-0.260 |
-0.415 |
| 299 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.259 |
0.437 |
| 300 |
244923_s_at |
ATMG01020
|
ORF153B |
-0.258 |
-0.078 |
| 301 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
-0.257 |
-0.307 |
| 302 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.256 |
-0.449 |
| 303 |
252976_s_at |
AT4G38550
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.256 |
0.016 |
| 304 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.255 |
0.034 |
| 305 |
257751_at |
AT3G18690
|
MKS1, MAP kinase substrate 1 |
-0.255 |
0.066 |
| 306 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
-0.253 |
0.107 |
| 307 |
250676_at |
AT5G06320
|
NHL3, NDR1/HIN1-like 3 |
-0.253 |
-0.263 |
| 308 |
251162_at |
AT3G63300
|
FKD1, FORKED 1 |
-0.252 |
-0.031 |
| 309 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
-0.252 |
-0.345 |
| 310 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.251 |
-0.202 |
| 311 |
250491_at |
AT5G09780
|
[Transcriptional factor B3 family protein] |
-0.251 |
-0.111 |
| 312 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
-0.251 |
-0.030 |
| 313 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.251 |
-0.225 |
| 314 |
247550_at |
AT5G61370
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.251 |
0.129 |
| 315 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.249 |
-0.488 |
| 316 |
265005_at |
AT1G61667
|
unknown |
-0.248 |
-0.023 |
| 317 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.248 |
-0.364 |
| 318 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.248 |
-0.473 |
| 319 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.247 |
-0.026 |
| 320 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.246 |
-0.495 |
| 321 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
-0.246 |
0.293 |