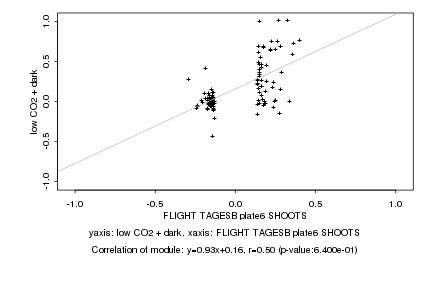
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
FLIGHT TAGESB plate6 SHOOTS |
Log2 signal ratio
low CO2 + dark |
| 1 |
263475_at |
AT2G31945
|
unknown |
0.397 |
0.768 |
| 2 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.359 |
0.727 |
| 3 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.353 |
0.592 |
| 4 |
264314_at |
AT1G70420
|
unknown |
0.332 |
0.009 |
| 5 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.319 |
1.018 |
| 6 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.283 |
0.366 |
| 7 |
246584_at |
AT5G14730
|
unknown |
0.281 |
0.158 |
| 8 |
254954_at |
AT4G10910
|
unknown |
0.277 |
0.697 |
| 9 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.274 |
-0.140 |
| 10 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.264 |
1.027 |
| 11 |
265387_at |
AT2G20670
|
unknown |
0.259 |
0.754 |
| 12 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.248 |
0.663 |
| 13 |
246552_at |
AT5G15420
|
unknown |
0.248 |
0.024 |
| 14 |
259566_at |
AT1G20520
|
unknown |
0.243 |
0.008 |
| 15 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.237 |
0.247 |
| 16 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.235 |
-0.073 |
| 17 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.231 |
0.176 |
| 18 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.223 |
0.759 |
| 19 |
248432_at |
AT5G51390
|
unknown |
0.217 |
0.644 |
| 20 |
255733_at |
AT1G25400
|
unknown |
0.215 |
0.663 |
| 21 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.188 |
0.253 |
| 22 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
0.188 |
0.452 |
| 23 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
0.185 |
-0.015 |
| 24 |
252072_at |
AT3G51710
|
[D-mannose binding lectin protein with Apple-like carbohydrate-binding domain] |
0.183 |
0.133 |
| 25 |
266196_at |
AT2G39110
|
[Protein kinase superfamily protein] |
0.180 |
0.004 |
| 26 |
251209_at |
AT3G62890
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.180 |
-0.017 |
| 27 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.173 |
0.680 |
| 28 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
0.171 |
-0.039 |
| 29 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.169 |
0.692 |
| 30 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
0.163 |
0.034 |
| 31 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.162 |
0.468 |
| 32 |
264379_at |
AT2G25200
|
unknown |
0.161 |
0.427 |
| 33 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.159 |
0.190 |
| 34 |
261558_at |
AT1G01770
|
unknown |
0.158 |
0.275 |
| 35 |
245831_at |
AT1G48840
|
unknown |
0.157 |
0.194 |
| 36 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
0.157 |
0.078 |
| 37 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.151 |
0.558 |
| 38 |
253997_at |
AT4G26090
|
RPS2, RESISTANT TO P. SYRINGAE 2 |
0.150 |
0.115 |
| 39 |
264238_at |
AT1G54740
|
unknown |
0.148 |
1.003 |
| 40 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.147 |
0.475 |
| 41 |
262127_at |
AT1G52550
|
unknown |
0.147 |
0.373 |
| 42 |
253364_at |
AT4G33160
|
[F-box family protein] |
0.147 |
-0.016 |
| 43 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.146 |
0.321 |
| 44 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.145 |
0.341 |
| 45 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.145 |
0.411 |
| 46 |
255836_at |
AT2G33440
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.143 |
0.013 |
| 47 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.143 |
0.492 |
| 48 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.142 |
0.169 |
| 49 |
259015_at |
AT3G07350
|
unknown |
0.141 |
0.616 |
| 50 |
266259_at |
AT2G27830
|
unknown |
0.139 |
0.691 |
| 51 |
254752_at |
AT4G13160
|
unknown |
0.137 |
0.236 |
| 52 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.137 |
0.276 |
| 53 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.136 |
-0.035 |
| 54 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.135 |
0.221 |
| 55 |
264550_at |
AT1G09450
|
AtHaspin, Haspin, Haspin-related gene |
0.135 |
-0.158 |
| 56 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.132 |
0.266 |
| 57 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.293 |
0.285 |
| 58 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.243 |
-0.087 |
| 59 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.241 |
-0.041 |
| 60 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.237 |
-0.051 |
| 61 |
245669_at |
AT1G28300
|
LEC2, LEAFY COTYLEDON 2 |
-0.216 |
0.022 |
| 62 |
262764_at |
AT1G13140
|
CYP86C3, cytochrome P450, family 86, subfamily C, polypeptide 3 |
-0.211 |
-0.012 |
| 63 |
266378_at |
AT2G14700
|
unknown |
-0.195 |
0.103 |
| 64 |
257562_at |
AT3G30480
|
unknown |
-0.192 |
0.046 |
| 65 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.188 |
0.416 |
| 66 |
251452_at |
AT3G60060
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.179 |
0.023 |
| 67 |
251748_at |
AT3G55680
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.179 |
0.043 |
| 68 |
260241_at |
AT1G63710
|
CYP86A7, cytochrome P450, family 86, subfamily A, polypeptide 7 |
-0.179 |
-0.091 |
| 69 |
250025_at |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
-0.176 |
-0.082 |
| 70 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.173 |
-0.018 |
| 71 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-0.171 |
-0.034 |
| 72 |
245700_at |
AT5G04180
|
ACA3, alpha carbonic anhydrase 3, ATACA3, ALPHA CARBONIC ANHYDRASE 3 |
-0.170 |
0.111 |
| 73 |
257028_at |
AT3G19230
|
[Leucine-rich repeat (LRR) family protein] |
-0.164 |
-0.009 |
| 74 |
265945_at |
AT2G19500
|
ATCKX2, CKX2, cytokinin oxidase 2 |
-0.164 |
0.051 |
| 75 |
250393_at |
AT5G10900
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.162 |
0.086 |
| 76 |
261807_at |
AT1G30515
|
unknown |
-0.162 |
-0.018 |
| 77 |
258358_at |
AT3G14380
|
[Uncharacterised protein family (UPF0497)] |
-0.160 |
-0.011 |
| 78 |
259714_at |
AT1G60980
|
ATGA20OX4, gibberellin 20-oxidase 4, GA20OX4, gibberellin 20-oxidase 4 |
-0.160 |
0.035 |
| 79 |
246896_at |
AT5G25550
|
[Leucine-rich repeat (LRR) family protein] |
-0.159 |
0.018 |
| 80 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.157 |
0.025 |
| 81 |
257517_at |
AT3G16330
|
unknown |
-0.156 |
-0.062 |
| 82 |
263324_at |
AT2G04220
|
unknown |
-0.156 |
-0.003 |
| 83 |
246754_at |
AT5G27910
|
NF-YC8, nuclear factor Y, subunit C8 |
-0.155 |
0.021 |
| 84 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.155 |
-0.062 |
| 85 |
255051_at |
AT4G09710
|
[non-LTR retrotransposon family (LINE), has a 4.7e-40 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
-0.154 |
0.034 |
| 86 |
266524_at |
AT2G16960
|
[ARM repeat superfamily protein] |
-0.152 |
-0.037 |
| 87 |
252307_at |
AT3G49280
|
unknown |
-0.152 |
0.053 |
| 88 |
266084_at |
AT2G37810
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.152 |
0.045 |
| 89 |
260860_at |
AT1G43810
|
unknown |
-0.152 |
-0.047 |
| 90 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.152 |
-0.044 |
| 91 |
257605_at |
AT3G13840
|
[GRAS family transcription factor] |
-0.152 |
0.025 |
| 92 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
-0.151 |
0.161 |
| 93 |
250616_at |
AT5G07280
|
EMS1, EXCESS MICROSPOROCYTES1, EXS, EXTRA SPOROGENOUS CELLS |
-0.151 |
-0.060 |
| 94 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.149 |
-0.035 |
| 95 |
257059_at |
AT3G15280
|
unknown |
-0.148 |
0.016 |
| 96 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.147 |
0.010 |
| 97 |
254469_at |
AT4G20470
|
unknown |
-0.146 |
0.087 |
| 98 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.145 |
-0.431 |
| 99 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.144 |
0.125 |
| 100 |
254631_at |
AT4G18610
|
LSH9, LIGHT SENSITIVE HYPOCOTYLS 9 |
-0.143 |
0.122 |
| 101 |
250574_at |
AT5G08230
|
[Tudor/PWWP/MBT domain-containing protein] |
-0.141 |
0.052 |
| 102 |
248054_at |
AT5G55820
|
WYR, WYRD |
-0.141 |
-0.091 |
| 103 |
263382_at |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
-0.141 |
-0.054 |
| 104 |
254972_at |
AT4G10440
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.140 |
0.001 |
| 105 |
263036_at |
AT1G23890
|
[NHL domain-containing protein] |
-0.140 |
0.044 |
| 106 |
257542_at |
AT3G26050
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.140 |
-0.108 |
| 107 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.140 |
-0.079 |
| 108 |
248939_at |
AT5G45790
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.140 |
0.010 |
| 109 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
-0.136 |
-0.021 |
| 110 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
-0.136 |
-0.207 |
| 111 |
260043_at |
AT1G41770
|
unknown |
-0.136 |
0.006 |