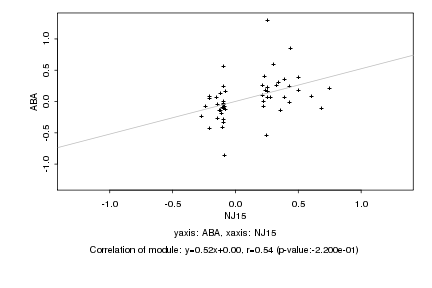
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
NJ15 |
Log2 signal ratio
ABA |
| 1 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.741 |
0.210 |
| 2 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.683 |
-0.096 |
| 3 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.605 |
0.089 |
| 4 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.497 |
0.396 |
| 5 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.496 |
0.182 |
| 6 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.434 |
0.861 |
| 7 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.428 |
0.253 |
| 8 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.427 |
-0.014 |
| 9 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.389 |
0.358 |
| 10 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.388 |
0.071 |
| 11 |
256518_at |
AT1G66080
|
unknown |
0.358 |
-0.135 |
| 12 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.336 |
0.315 |
| 13 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.326 |
0.270 |
| 14 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.301 |
0.597 |
| 15 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.274 |
0.074 |
| 16 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.254 |
1.310 |
| 17 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.254 |
0.168 |
| 18 |
247509_at |
AT5G62020
|
AT-HSFB2A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR B2A, HSFB2A, heat shock transcription factor B2A |
0.252 |
0.228 |
| 19 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
0.250 |
0.068 |
| 20 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.250 |
0.167 |
| 21 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.241 |
-0.538 |
| 22 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
0.234 |
0.188 |
| 23 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
0.230 |
0.404 |
| 24 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.223 |
-0.074 |
| 25 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.218 |
0.015 |
| 26 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.209 |
0.099 |
| 27 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.208 |
0.262 |
| 28 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.275 |
-0.233 |
| 29 |
254459_at |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
-0.245 |
-0.067 |
| 30 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.214 |
-0.430 |
| 31 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.212 |
0.084 |
| 32 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.208 |
0.054 |
| 33 |
257244_at |
AT3G24240
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
-0.157 |
0.067 |
| 34 |
265611_at |
AT2G25510
|
unknown |
-0.146 |
-0.045 |
| 35 |
263596_at |
AT2G01900
|
[DNAse I-like superfamily protein] |
-0.146 |
-0.263 |
| 36 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
-0.129 |
-0.143 |
| 37 |
252237_at |
AT3G49950
|
[GRAS family transcription factor] |
-0.126 |
0.143 |
| 38 |
260617_at |
AT1G53345
|
unknown |
-0.120 |
-0.133 |
| 39 |
254408_at |
AT4G21390
|
B120 |
-0.113 |
-0.182 |
| 40 |
251822_at |
AT3G55060
|
unknown |
-0.110 |
-0.090 |
| 41 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.109 |
-0.406 |
| 42 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.102 |
-0.042 |
| 43 |
247832_at |
AT5G58550
|
EOL2, ETO1-like 2 |
-0.102 |
0.009 |
| 44 |
250231_at |
AT5G13910
|
LEP, LEAFY PETIOLE |
-0.100 |
-0.108 |
| 45 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.100 |
-0.016 |
| 46 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.098 |
-0.281 |
| 47 |
257314_at |
AT3G26590
|
[MATE efflux family protein] |
-0.097 |
-0.327 |
| 48 |
255413_at |
AT4G03140
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.096 |
0.249 |
| 49 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.096 |
0.567 |
| 50 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.091 |
-0.860 |
| 51 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.088 |
-0.077 |
| 52 |
257554_at |
AT3G24890
|
ATVAMP728, VAMP728, vesicle-associated membrane protein 728 |
-0.087 |
-0.125 |
| 53 |
254301_at |
AT4G22790
|
[MATE efflux family protein] |
-0.084 |
0.173 |