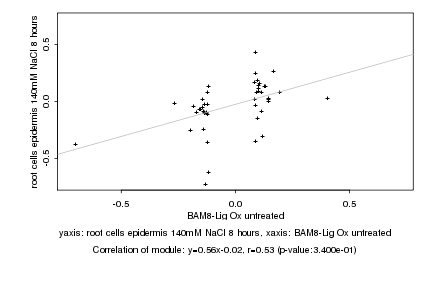
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
BAM8-Lig Ox untreated |
Log2 signal ratio
root cells epidermis 140mM NaCl 8 hours |
| 1 |
248997_at |
AT5G45300
|
BAM8, BETA-AMYLASE 8, BMY2, beta-amylase 2 |
0.400 |
0.029 |
| 2 |
258374_at |
AT3G14360
|
[alpha/beta-Hydrolases superfamily protein] |
0.192 |
0.088 |
| 3 |
251637_at |
AT3G57570
|
[ARM repeat superfamily protein] |
0.164 |
0.264 |
| 4 |
261863_at |
AT1G50630
|
unknown |
0.142 |
0.001 |
| 5 |
256998_at |
AT3G14180
|
ASIL2, Arabidopsis 6B-interacting protein 1-like 2 |
0.142 |
0.031 |
| 6 |
249069_at |
AT5G44010
|
unknown |
0.141 |
0.020 |
| 7 |
267065_at |
AT2G41080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.129 |
0.134 |
| 8 |
262150_at |
AT1G52520
|
FRS6, FAR1-related sequence 6 |
0.123 |
0.133 |
| 9 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
0.115 |
-0.305 |
| 10 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
0.114 |
0.085 |
| 11 |
252352_at |
AT3G48185
|
unknown |
0.113 |
-0.085 |
| 12 |
255485_at |
AT4G02550
|
unknown |
0.105 |
0.164 |
| 13 |
263681_at |
AT1G26840
|
ATORC6, ARABIDOPSIS THALIANA ORIGIN RECOGNITION COMPLEX PROTEIN 6, ORC6, origin recognition complex protein 6 |
0.100 |
0.141 |
| 14 |
254054_at |
AT4G25320
|
AHL3, AT-HOOK MOTIF NUCLEAR LOCALIZED PROTEIN 3 |
0.099 |
0.116 |
| 15 |
256409_at |
AT1G66620
|
[Protein with RING/U-box and TRAF-like domains] |
0.097 |
0.093 |
| 16 |
257725_at |
AT3G18524
|
ATMSH2, MSH2, MUTS homolog 2 |
0.095 |
0.185 |
| 17 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.093 |
-0.143 |
| 18 |
256016_at |
AT1G19240
|
unknown |
0.092 |
0.082 |
| 19 |
261916_at |
AT1G65910
|
anac028, NAC domain containing protein 28, NAC028, NAC domain containing protein 28 |
0.087 |
-0.033 |
| 20 |
258652_at |
AT3G09910
|
ATRAB18C, ATRABC2B, RAB GTPase homolog C2B, RABC2b, RAB GTPase homolog C2B |
0.087 |
0.432 |
| 21 |
256384_at |
AT1G66660
|
[Protein with RING/U-box and TRAF-like domains] |
0.084 |
0.252 |
| 22 |
255175_at |
AT4G07960
|
ATCSLC12, Cellulose-synthase-like C12, CSLC12, CELLULOSE-SYNTHASE LIKE C12, CSLC12, Cellulose-synthase-like C12 |
0.084 |
-0.347 |
| 23 |
262410_at |
AT1G34770
|
unknown |
0.083 |
0.175 |
| 24 |
262500_at |
AT1G21760
|
ATFBP7, F-BOX PROTEIN 7, FBP7, F-box protein 7 |
0.081 |
0.026 |
| 25 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.704 |
-0.373 |
| 26 |
267259_at |
AT2G23090
|
[Uncharacterised protein family SERF] |
-0.269 |
-0.009 |
| 27 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.199 |
-0.253 |
| 28 |
257584_at |
AT1G54955
|
unknown |
-0.185 |
-0.035 |
| 29 |
265093_at |
AT1G03905
|
ABCI19, ATP-binding cassette I19 |
-0.172 |
-0.095 |
| 30 |
256180_at |
AT1G51810
|
[Leucine-rich repeat protein kinase family protein] |
-0.161 |
-0.067 |
| 31 |
255770_at |
AT1G18560
|
[BED zinc finger ] |
-0.157 |
-0.068 |
| 32 |
267098_at |
AT2G41451
|
unknown |
-0.148 |
0.020 |
| 33 |
265620_at |
AT2G27310
|
[F-box family protein] |
-0.145 |
-0.048 |
| 34 |
264495_at |
AT1G27380
|
RIC2, ROP-interactive CRIB motif-containing protein 2 |
-0.144 |
-0.245 |
| 35 |
250631_at |
AT5G07430
|
[Pectin lyase-like superfamily protein] |
-0.141 |
-0.085 |
| 36 |
249987_at |
AT5G18490
|
unknown |
-0.139 |
-0.099 |
| 37 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.138 |
-0.089 |
| 38 |
262465_at |
AT1G50270
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.136 |
-0.024 |
| 39 |
256847_at |
AT3G27950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.132 |
-0.721 |
| 40 |
262615_at |
AT1G13950
|
ATELF5A-1, EUKARYOTIC ELONGATION FACTOR 5A-1, EIF-5A, EIF5A, EUKARYOTIC ELONGATION FACTOR 5A, ELF5A-1, eukaryotic elongation factor 5A-1 |
-0.131 |
-0.095 |
| 41 |
252675_at |
AT3G44270
|
[copia-like retrotransposon family, has a 1.1e-26 P-value blast match to gb|AAG52950.1| putative envelope protein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
-0.131 |
-0.100 |
| 42 |
250697_at |
AT5G06800
|
[myb-like HTH transcriptional regulator family protein] |
-0.126 |
-0.359 |
| 43 |
255705_at |
AT4G00160
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.126 |
-0.023 |
| 44 |
250746_at |
AT5G05880
|
[UDP-Glycosyltransferase superfamily protein] |
-0.126 |
0.086 |
| 45 |
258841_at |
AT3G04660
|
[F-box and associated interaction domains-containing protein] |
-0.125 |
-0.107 |
| 46 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.121 |
-0.621 |
| 47 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.120 |
0.138 |