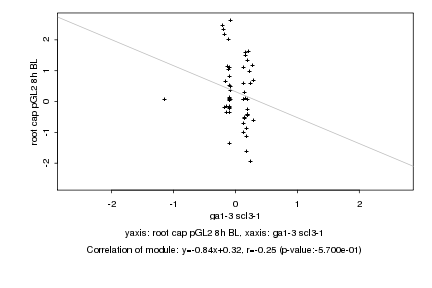
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ga1-3 scl3-1 |
Log2 signal ratio
root cap pGL2 8h BL |
| 1 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.290 |
0.690 |
| 2 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.276 |
-0.597 |
| 3 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.262 |
1.180 |
| 4 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.238 |
-1.919 |
| 5 |
248153_at |
AT5G54250
|
ATCNGC4, cyclic nucleotide-gated cation channel 4, CNGC4, cyclic nucleotide-gated cation channel 4, DND2, DEFENSE, NO DEATH 2, HLM1 |
0.232 |
0.611 |
| 6 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.211 |
0.999 |
| 7 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.202 |
1.630 |
| 8 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.187 |
-0.248 |
| 9 |
264636_at |
AT1G65490
|
unknown |
0.186 |
0.079 |
| 10 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
0.183 |
-0.442 |
| 11 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
0.182 |
-0.414 |
| 12 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.179 |
-0.406 |
| 13 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.179 |
1.358 |
| 14 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.172 |
-0.858 |
| 15 |
250620_at |
AT5G07190
|
ATS3, seed gene 3 |
0.172 |
-1.612 |
| 16 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.163 |
-1.130 |
| 17 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
0.158 |
1.505 |
| 18 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.155 |
0.101 |
| 19 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.150 |
1.594 |
| 20 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.141 |
-0.540 |
| 21 |
263787_at |
AT2G46420
|
unknown |
0.139 |
0.306 |
| 22 |
255720_at |
AT1G32060
|
PRK, phosphoribulokinase |
0.133 |
-0.491 |
| 23 |
248674_at |
AT5G48800
|
[Phototropic-responsive NPH3 family protein] |
0.127 |
-0.996 |
| 24 |
264635_at |
AT1G65500
|
unknown |
0.127 |
-0.688 |
| 25 |
248593_at |
AT5G49180
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.125 |
0.079 |
| 26 |
267167_at |
AT2G37740
|
ATZFP10, ZINC-FINGER PROTEIN 10, ZFP10, zinc-finger protein 10 |
0.125 |
0.079 |
| 27 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
0.124 |
0.592 |
| 28 |
261522_at |
AT1G71710
|
[DNAse I-like superfamily protein] |
0.123 |
1.105 |
| 29 |
261866_at |
AT1G50420
|
SCL-3, SCARECROW-LIKE 3, SCL3, scarecrow-like 3 |
-1.152 |
0.078 |
| 30 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.222 |
2.481 |
| 31 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.208 |
2.358 |
| 32 |
260668_at |
AT1G19530
|
unknown |
-0.192 |
2.191 |
| 33 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.184 |
-0.163 |
| 34 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.162 |
0.677 |
| 35 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
-0.148 |
-0.341 |
| 36 |
249127_at |
AT5G43500
|
ARP9, actin-related protein 9, ATARP9, actin-related protein 9 |
-0.147 |
-0.132 |
| 37 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.136 |
1.163 |
| 38 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.122 |
2.012 |
| 39 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.119 |
1.053 |
| 40 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.111 |
-0.354 |
| 41 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
-0.110 |
0.101 |
| 42 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.110 |
0.839 |
| 43 |
252153_at |
AT3G51360
|
[Eukaryotic aspartyl protease family protein] |
-0.110 |
-0.159 |
| 44 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.108 |
-0.195 |
| 45 |
256520_at |
AT1G66030
|
pseudogene |
-0.107 |
0.092 |
| 46 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
-0.104 |
0.064 |
| 47 |
267638_at |
AT2G42210
|
ATOEP16-3, OEP16-3 |
-0.104 |
0.139 |
| 48 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.103 |
-1.357 |
| 49 |
255062_at |
AT4G08940
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.102 |
-0.181 |
| 50 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
-0.097 |
1.104 |
| 51 |
249824_at |
AT5G23380
|
unknown |
-0.096 |
0.529 |
| 52 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
-0.093 |
0.497 |
| 53 |
251660_at |
AT3G57160
|
unknown |
-0.092 |
0.367 |
| 54 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
-0.086 |
0.079 |
| 55 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.086 |
2.659 |