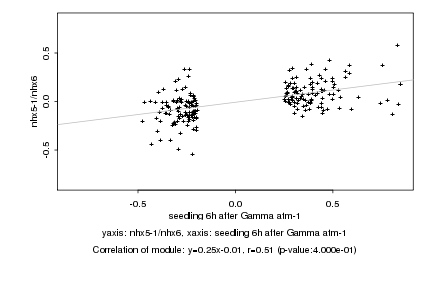
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
seedling 6h after Gamma atm-1 |
Log2 signal ratio
nhx5-1/nhx6 |
| 1 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
0.845 |
0.176 |
| 2 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
0.844 |
0.183 |
| 3 |
262211_at |
AT1G74930
|
ORA47 |
0.832 |
-0.023 |
| 4 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.828 |
0.586 |
| 5 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
0.802 |
-0.124 |
| 6 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
0.777 |
0.018 |
| 7 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.750 |
0.380 |
| 8 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.741 |
-0.016 |
| 9 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.630 |
0.047 |
| 10 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.591 |
-0.076 |
| 11 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.582 |
0.376 |
| 12 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.582 |
0.296 |
| 13 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
0.561 |
0.314 |
| 14 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.561 |
0.255 |
| 15 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.538 |
0.044 |
| 16 |
254287_at |
AT4G22960
|
unknown |
0.533 |
-0.068 |
| 17 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.525 |
0.117 |
| 18 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.506 |
0.183 |
| 19 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
0.503 |
0.150 |
| 20 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
0.501 |
0.077 |
| 21 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.496 |
0.211 |
| 22 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
0.494 |
0.031 |
| 23 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.494 |
0.239 |
| 24 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.481 |
0.426 |
| 25 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.481 |
0.081 |
| 26 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.470 |
-0.076 |
| 27 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.461 |
0.335 |
| 28 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.460 |
0.213 |
| 29 |
245041_at |
AT2G26530
|
AR781 |
0.456 |
0.123 |
| 30 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.449 |
-0.086 |
| 31 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.446 |
0.039 |
| 32 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.445 |
-0.119 |
| 33 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
0.442 |
0.110 |
| 34 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
0.441 |
-0.014 |
| 35 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.441 |
0.112 |
| 36 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
0.440 |
-0.060 |
| 37 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.438 |
0.128 |
| 38 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.437 |
0.246 |
| 39 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.431 |
0.269 |
| 40 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
0.423 |
-0.061 |
| 41 |
246791_at |
AT5G27280
|
[Zim17-type zinc finger protein] |
0.417 |
0.196 |
| 42 |
251372_at |
AT3G60520
|
unknown |
0.416 |
0.092 |
| 43 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.410 |
0.065 |
| 44 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
0.395 |
0.199 |
| 45 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.395 |
0.087 |
| 46 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.393 |
0.150 |
| 47 |
249941_at |
AT5G22270
|
unknown |
0.391 |
0.132 |
| 48 |
262301_at |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.390 |
0.016 |
| 49 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.388 |
0.027 |
| 50 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.386 |
0.383 |
| 51 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
0.385 |
0.178 |
| 52 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.385 |
-0.017 |
| 53 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.383 |
0.000 |
| 54 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
0.382 |
0.238 |
| 55 |
252661_at |
AT3G44450
|
unknown |
0.380 |
-0.077 |
| 56 |
245600_at |
AT4G14230
|
unknown |
0.374 |
0.080 |
| 57 |
266500_at |
AT2G06925
|
ATSPLA2-ALPHA, PHOSPHOLIPASE A2-ALPHA, PLA2-ALPHA |
0.371 |
-0.001 |
| 58 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
0.364 |
0.332 |
| 59 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.364 |
-0.036 |
| 60 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.359 |
-0.091 |
| 61 |
255953_at |
AT1G22070
|
TGA3, TGA1A-related gene 3 |
0.355 |
0.020 |
| 62 |
245094_at |
AT2G40840
|
DPE2, disproportionating enzyme 2 |
0.353 |
0.148 |
| 63 |
247498_at |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
0.345 |
0.021 |
| 64 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
0.345 |
-0.045 |
| 65 |
254662_at |
AT4G18270
|
ATTRANS11, ARABIDOPSIS THALIANA TRANSLOCASE 11, TRANS11, translocase 11 |
0.344 |
0.074 |
| 66 |
263515_at |
AT2G21640
|
unknown |
0.339 |
-0.145 |
| 67 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.336 |
0.055 |
| 68 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.333 |
0.039 |
| 69 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.331 |
0.121 |
| 70 |
259224_at |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
0.319 |
0.019 |
| 71 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.317 |
-0.013 |
| 72 |
266831_at |
AT2G22830
|
SQE2, squalene epoxidase 2 |
0.317 |
0.088 |
| 73 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.315 |
-0.075 |
| 74 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.312 |
0.147 |
| 75 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
0.311 |
0.041 |
| 76 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
0.310 |
0.106 |
| 77 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.310 |
0.251 |
| 78 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.304 |
0.106 |
| 79 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
0.302 |
-0.042 |
| 80 |
247450_at |
AT5G62350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.301 |
0.095 |
| 81 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.299 |
-0.117 |
| 82 |
261661_at |
AT1G18360
|
[alpha/beta-Hydrolases superfamily protein] |
0.298 |
-0.001 |
| 83 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
0.298 |
0.189 |
| 84 |
250382_at |
AT5G11580
|
[Regulator of chromosome condensation (RCC1) family protein] |
0.294 |
0.137 |
| 85 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.292 |
0.062 |
| 86 |
265481_at |
AT2G15960
|
unknown |
0.290 |
0.342 |
| 87 |
266396_at |
AT2G38790
|
unknown |
0.289 |
0.018 |
| 88 |
262609_at |
AT1G13930
|
unknown |
0.289 |
0.047 |
| 89 |
260636_at |
AT1G62430
|
ATCDS1, CDP-diacylglycerol synthase 1, CDS1, CDP-diacylglycerol synthase 1 |
0.289 |
0.136 |
| 90 |
262236_at |
AT1G48330
|
unknown |
0.289 |
0.247 |
| 91 |
250005_at |
AT5G18760
|
[RING/U-box superfamily protein] |
0.287 |
0.045 |
| 92 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.283 |
0.024 |
| 93 |
255585_at |
AT4G01550
|
ANAC069, NAC domain containing protein 69, NAC069, NAC domain containing protein 69, NTM2, NAC with Transmembrane Motif 2 |
0.281 |
0.196 |
| 94 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
0.279 |
-0.021 |
| 95 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
0.275 |
0.320 |
| 96 |
249355_at |
AT5G40500
|
unknown |
0.275 |
-0.004 |
| 97 |
263249_at |
AT2G31360
|
ADS2, 16:0delta9 desaturase 2, AtADS2 |
0.273 |
-0.001 |
| 98 |
252770_at |
AT3G42860
|
[zinc knuckle (CCHC-type) family protein] |
0.272 |
-0.000 |
| 99 |
247239_at |
AT5G64640
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.268 |
0.030 |
| 100 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
0.268 |
0.171 |
| 101 |
262784_at |
AT1G10760
|
GWD, GWD1, SEX1, STARCH EXCESS 1, SOP, SOP1 |
0.268 |
0.156 |
| 102 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
0.267 |
0.099 |
| 103 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.264 |
0.083 |
| 104 |
245563_at |
AT4G14580
|
CIPK4, CBL-interacting protein kinase 4, SnRK3.3, SNF1-RELATED PROTEIN KINASE 3.3 |
0.262 |
0.138 |
| 105 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.261 |
0.067 |
| 106 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.258 |
0.085 |
| 107 |
256091_at |
AT1G20693
|
HMG BETA 1, HIGH MOBILITY GROUP BETA 1, HMGB2, high mobility group B2, NFD02, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 02, NFD2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 2 |
0.256 |
0.058 |
| 108 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.255 |
0.008 |
| 109 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
0.254 |
0.205 |
| 110 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.251 |
0.021 |
| 111 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.479 |
-0.201 |
| 112 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
-0.471 |
-0.003 |
| 113 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.439 |
0.002 |
| 114 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.432 |
-0.435 |
| 115 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
-0.411 |
-0.010 |
| 116 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.409 |
-0.168 |
| 117 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.405 |
-0.305 |
| 118 |
246552_at |
AT5G15420
|
unknown |
-0.396 |
0.101 |
| 119 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.390 |
-0.106 |
| 120 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.389 |
-0.397 |
| 121 |
253919_at |
AT4G27350
|
unknown |
-0.387 |
-0.196 |
| 122 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.378 |
-0.070 |
| 123 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.376 |
0.000 |
| 124 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
-0.374 |
0.124 |
| 125 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.361 |
-0.120 |
| 126 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.354 |
-0.037 |
| 127 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.354 |
-0.120 |
| 128 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.354 |
-0.000 |
| 129 |
261382_at |
AT1G05470
|
CVP2, COTYLEDON VASCULAR PATTERN 2 |
-0.348 |
-0.051 |
| 130 |
248057_at |
AT5G55520
|
unknown |
-0.344 |
-0.058 |
| 131 |
253185_at |
AT4G35240
|
unknown |
-0.339 |
-0.125 |
| 132 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
-0.338 |
-0.092 |
| 133 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.338 |
-0.084 |
| 134 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.336 |
-0.393 |
| 135 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.325 |
-0.247 |
| 136 |
252882_at |
AT4G39675
|
unknown |
-0.321 |
0.016 |
| 137 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.319 |
-0.221 |
| 138 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.319 |
0.006 |
| 139 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.317 |
-0.210 |
| 140 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.312 |
-0.230 |
| 141 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
-0.310 |
0.215 |
| 142 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.306 |
-0.004 |
| 143 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
-0.304 |
0.122 |
| 144 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.302 |
-0.064 |
| 145 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-0.300 |
-0.077 |
| 146 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.300 |
0.019 |
| 147 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.299 |
-0.204 |
| 148 |
266222_at |
AT2G28780
|
unknown |
-0.297 |
0.234 |
| 149 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.296 |
-0.099 |
| 150 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.295 |
-0.152 |
| 151 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
-0.294 |
-0.494 |
| 152 |
258545_at |
AT3G07050
|
NSN1, nucleostemin-like 1 |
-0.290 |
-0.166 |
| 153 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.290 |
-0.058 |
| 154 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
-0.290 |
0.036 |
| 155 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.287 |
-0.321 |
| 156 |
258209_at |
AT3G14060
|
unknown |
-0.287 |
0.005 |
| 157 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.283 |
-0.124 |
| 158 |
248295_at |
AT5G53070
|
[Ribosomal protein L9/RNase H1] |
-0.281 |
0.030 |
| 159 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.279 |
-0.009 |
| 160 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.276 |
0.128 |
| 161 |
246120_at |
AT5G20360
|
Phox3, Phox3 |
-0.274 |
-0.147 |
| 162 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
-0.267 |
-0.196 |
| 163 |
258362_at |
AT3G14280
|
unknown |
-0.264 |
0.335 |
| 164 |
263330_at |
AT2G15320
|
[Leucine-rich repeat (LRR) family protein] |
-0.260 |
-0.142 |
| 165 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.259 |
0.145 |
| 166 |
246926_at |
AT5G25240
|
unknown |
-0.258 |
-0.033 |
| 167 |
264636_at |
AT1G65490
|
unknown |
-0.257 |
-0.109 |
| 168 |
249951_at |
AT5G18930
|
BUD2, BUSHY AND DWARF 2, SAMDC4 |
-0.254 |
0.002 |
| 169 |
256916_at |
AT3G24050
|
GATA1, GATA transcription factor 1 |
-0.254 |
-0.041 |
| 170 |
248691_at |
AT5G48310
|
unknown |
-0.253 |
-0.132 |
| 171 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.250 |
-0.245 |
| 172 |
253243_at |
AT4G34560
|
unknown |
-0.247 |
-0.144 |
| 173 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.243 |
-0.159 |
| 174 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.243 |
-0.202 |
| 175 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.242 |
0.266 |
| 176 |
266424_at |
AT2G41330
|
[Glutaredoxin family protein] |
-0.242 |
0.006 |
| 177 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
-0.242 |
-0.006 |
| 178 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.240 |
-0.183 |
| 179 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
-0.238 |
0.338 |
| 180 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.237 |
-0.061 |
| 181 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.236 |
-0.011 |
| 182 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.236 |
-0.142 |
| 183 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.236 |
-0.111 |
| 184 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.233 |
0.063 |
| 185 |
248772_at |
AT5G47800
|
[Phototropic-responsive NPH3 family protein] |
-0.232 |
0.087 |
| 186 |
247029_at |
AT5G67190
|
DEAR2, DREB and EAR motif protein 2 |
-0.228 |
-0.037 |
| 187 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
-0.228 |
-0.141 |
| 188 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.227 |
-0.003 |
| 189 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.225 |
-0.033 |
| 190 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.223 |
-0.139 |
| 191 |
247014_at |
AT5G67630
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.222 |
-0.124 |
| 192 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.222 |
-0.540 |
| 193 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.221 |
-0.161 |
| 194 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.221 |
-0.106 |
| 195 |
247605_at |
AT5G60970
|
TCP5, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 5 |
-0.219 |
-0.098 |
| 196 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
-0.219 |
0.070 |
| 197 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.219 |
0.005 |
| 198 |
265431_at |
AT2G20680
|
AtMAN2, MAN2, endo-beta-mannase 2 |
-0.218 |
0.054 |
| 199 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.217 |
-0.042 |
| 200 |
257076_at |
AT3G19680
|
unknown |
-0.217 |
-0.284 |
| 201 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-0.216 |
-0.188 |
| 202 |
257838_at |
AT3G25210
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.216 |
0.061 |
| 203 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
-0.215 |
0.039 |
| 204 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
-0.213 |
-0.123 |
| 205 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
-0.213 |
-0.155 |
| 206 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.213 |
-0.097 |
| 207 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
-0.212 |
0.017 |
| 208 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
-0.210 |
-0.121 |
| 209 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
-0.207 |
0.015 |
| 210 |
251371_at |
AT3G60360
|
EDA14, EMBRYO SAC DEVELOPMENT ARREST 14, UTP11, U3 SMALL NUCLEOLAR RNA-ASSOCIATED PROTEIN 11 |
-0.206 |
-0.094 |
| 211 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.205 |
-0.093 |
| 212 |
245090_at |
AT2G40900
|
UMAMIT11, Usually multiple acids move in and out Transporters 11 |
-0.205 |
-0.047 |
| 213 |
259823_at |
AT1G66250
|
[O-Glycosyl hydrolases family 17 protein] |
-0.204 |
-0.164 |
| 214 |
259422_at |
AT1G13810
|
[Restriction endonuclease, type II-like superfamily protein] |
-0.203 |
-0.014 |
| 215 |
251059_at |
AT5G01810
|
ATPK10, PROTEIN KINASE 10, CIPK15, CBL-interacting protein kinase 15, PKS3, SIP2, SOS3-INTERACTING PROTEIN 2, SNRK3.1, SNF1-RELATED PROTEIN KINASE 3.1 |
-0.203 |
-0.173 |
| 216 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.203 |
-0.295 |
| 217 |
259228_at |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.202 |
-0.036 |
| 218 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.201 |
-0.267 |
| 219 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
-0.199 |
-0.086 |