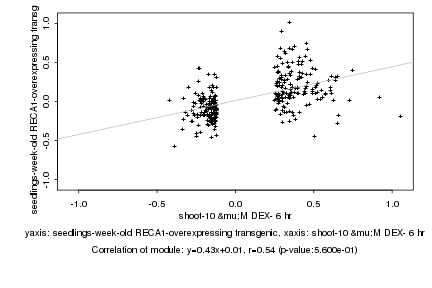
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
shoot-10 μM DEX- 6 hr |
Log2 signal ratio
seedlings-week-old RECA1-overexpressing transgenic |
| 1 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
1.051 |
-0.180 |
| 2 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.918 |
0.058 |
| 3 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.744 |
0.404 |
| 4 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.723 |
0.017 |
| 5 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.652 |
-0.168 |
| 6 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.650 |
0.325 |
| 7 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.649 |
-0.276 |
| 8 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.638 |
0.308 |
| 9 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.634 |
0.270 |
| 10 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.620 |
0.021 |
| 11 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.611 |
0.107 |
| 12 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.611 |
0.330 |
| 13 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.602 |
0.146 |
| 14 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.602 |
0.185 |
| 15 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.593 |
0.272 |
| 16 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.572 |
0.104 |
| 17 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.570 |
0.101 |
| 18 |
265387_at |
AT2G20670
|
unknown |
0.555 |
0.062 |
| 19 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.546 |
0.149 |
| 20 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.539 |
0.030 |
| 21 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.518 |
0.037 |
| 22 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.518 |
0.119 |
| 23 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.518 |
0.231 |
| 24 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.514 |
0.214 |
| 25 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.510 |
0.109 |
| 26 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.510 |
0.417 |
| 27 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.506 |
0.187 |
| 28 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.504 |
-0.438 |
| 29 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.491 |
0.155 |
| 30 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.491 |
0.113 |
| 31 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.488 |
0.427 |
| 32 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.488 |
0.136 |
| 33 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.478 |
0.357 |
| 34 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.473 |
0.527 |
| 35 |
254954_at |
AT4G10910
|
unknown |
0.471 |
-0.030 |
| 36 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.457 |
0.256 |
| 37 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.457 |
0.669 |
| 38 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.453 |
0.202 |
| 39 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.453 |
0.753 |
| 40 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.451 |
-0.047 |
| 41 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.450 |
0.349 |
| 42 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.444 |
0.105 |
| 43 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.443 |
0.584 |
| 44 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.441 |
0.170 |
| 45 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.435 |
0.140 |
| 46 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.429 |
0.098 |
| 47 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.422 |
0.363 |
| 48 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.422 |
0.519 |
| 49 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.419 |
0.487 |
| 50 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.417 |
0.320 |
| 51 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.408 |
0.567 |
| 52 |
245662_at |
AT1G28190
|
unknown |
0.407 |
0.303 |
| 53 |
266658_at |
AT2G25735
|
unknown |
0.406 |
0.383 |
| 54 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.404 |
-0.137 |
| 55 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.404 |
0.321 |
| 56 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.399 |
0.481 |
| 57 |
256442_at |
AT3G10930
|
unknown |
0.399 |
0.516 |
| 58 |
264379_at |
AT2G25200
|
unknown |
0.398 |
0.105 |
| 59 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.393 |
0.173 |
| 60 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.393 |
0.105 |
| 61 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.392 |
0.294 |
| 62 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.390 |
0.191 |
| 63 |
260744_at |
AT1G15010
|
unknown |
0.382 |
0.169 |
| 64 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.377 |
-0.223 |
| 65 |
245041_at |
AT2G26530
|
AR781 |
0.375 |
0.123 |
| 66 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.374 |
0.715 |
| 67 |
263799_at |
AT2G24550
|
unknown |
0.369 |
0.054 |
| 68 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.368 |
-0.167 |
| 69 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.362 |
0.670 |
| 70 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
0.362 |
-0.131 |
| 71 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.362 |
0.507 |
| 72 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.361 |
0.183 |
| 73 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.358 |
0.192 |
| 74 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.357 |
0.127 |
| 75 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.352 |
0.394 |
| 76 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
0.351 |
0.219 |
| 77 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.351 |
0.223 |
| 78 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.350 |
0.157 |
| 79 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.347 |
0.107 |
| 80 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.347 |
0.342 |
| 81 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.346 |
0.079 |
| 82 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.345 |
0.039 |
| 83 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.344 |
1.024 |
| 84 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.343 |
-0.249 |
| 85 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.342 |
0.091 |
| 86 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.341 |
0.687 |
| 87 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.341 |
0.082 |
| 88 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.341 |
0.088 |
| 89 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.341 |
0.370 |
| 90 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.339 |
-0.124 |
| 91 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
0.339 |
0.039 |
| 92 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.338 |
0.455 |
| 93 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.335 |
0.059 |
| 94 |
265276_at |
AT2G28400
|
unknown |
0.335 |
0.444 |
| 95 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.334 |
0.059 |
| 96 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.331 |
0.287 |
| 97 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.328 |
-0.024 |
| 98 |
264580_at |
AT1G05340
|
unknown |
0.328 |
0.511 |
| 99 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.324 |
0.115 |
| 100 |
245341_at |
AT4G16447
|
unknown |
0.323 |
-0.130 |
| 101 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.319 |
0.345 |
| 102 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.319 |
0.623 |
| 103 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.317 |
0.052 |
| 104 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
0.314 |
0.173 |
| 105 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.313 |
0.642 |
| 106 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.312 |
0.251 |
| 107 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.310 |
-0.070 |
| 108 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.309 |
0.281 |
| 109 |
247754_at |
AT5G59080
|
unknown |
0.307 |
-0.133 |
| 110 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
0.304 |
-0.062 |
| 111 |
261567_at |
AT1G33055
|
unknown |
0.302 |
0.128 |
| 112 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.300 |
-0.108 |
| 113 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.299 |
-0.265 |
| 114 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.298 |
0.141 |
| 115 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.297 |
0.004 |
| 116 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.295 |
0.489 |
| 117 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.294 |
0.041 |
| 118 |
265648_at |
AT2G27500
|
[Glycosyl hydrolase superfamily protein] |
0.294 |
0.209 |
| 119 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.294 |
0.096 |
| 120 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.293 |
0.906 |
| 121 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.293 |
0.337 |
| 122 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.292 |
0.180 |
| 123 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
0.291 |
0.063 |
| 124 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
0.291 |
0.315 |
| 125 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.290 |
0.289 |
| 126 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.286 |
0.682 |
| 127 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.284 |
-0.161 |
| 128 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.283 |
0.558 |
| 129 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.281 |
0.211 |
| 130 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.280 |
0.079 |
| 131 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.280 |
0.001 |
| 132 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.278 |
0.304 |
| 133 |
258225_at |
AT3G15630
|
unknown |
0.278 |
0.025 |
| 134 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.277 |
0.250 |
| 135 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.275 |
0.017 |
| 136 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.272 |
0.375 |
| 137 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.271 |
0.103 |
| 138 |
255527_at |
AT4G02360
|
unknown |
0.270 |
-0.015 |
| 139 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.270 |
0.396 |
| 140 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.270 |
0.005 |
| 141 |
253814_at |
AT4G28290
|
unknown |
0.269 |
0.044 |
| 142 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.268 |
0.580 |
| 143 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.268 |
0.044 |
| 144 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.267 |
0.459 |
| 145 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.267 |
0.083 |
| 146 |
253643_at |
AT4G29780
|
unknown |
0.265 |
0.262 |
| 147 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.264 |
0.380 |
| 148 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.263 |
0.199 |
| 149 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
0.263 |
0.266 |
| 150 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.263 |
0.068 |
| 151 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
0.261 |
0.234 |
| 152 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.261 |
0.101 |
| 153 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.257 |
0.213 |
| 154 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.256 |
-0.101 |
| 155 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.255 |
0.035 |
| 156 |
253322_at |
AT4G33980
|
unknown |
0.252 |
0.110 |
| 157 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.252 |
-0.108 |
| 158 |
259479_at |
AT1G19020
|
unknown |
0.249 |
0.445 |
| 159 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.248 |
0.023 |
| 160 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-0.427 |
0.017 |
| 161 |
265892_at |
AT2G15020
|
unknown |
-0.395 |
-0.568 |
| 162 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.338 |
-0.349 |
| 163 |
254482_at |
AT4G20370
|
TSF, TWIN SISTER OF FT |
-0.337 |
0.042 |
| 164 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.332 |
-0.223 |
| 165 |
253155_at |
AT4G35720
|
unknown |
-0.324 |
-0.134 |
| 166 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.311 |
-0.170 |
| 167 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
-0.303 |
0.180 |
| 168 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.282 |
-0.103 |
| 169 |
252661_at |
AT3G44450
|
unknown |
-0.281 |
-0.250 |
| 170 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.280 |
-0.250 |
| 171 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
-0.274 |
-0.063 |
| 172 |
251223_at |
AT3G62610
|
ATMYB11, myb domain protein 11, MYB11, myb domain protein 11, PFG2, PRODUCTION OF FLAVONOL GLYCOSIDES 2 |
-0.272 |
-0.008 |
| 173 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.269 |
-0.144 |
| 174 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.263 |
-0.170 |
| 175 |
259574_at |
AT1G35310
|
MLP168, MLP-like protein 168 |
-0.260 |
0.113 |
| 176 |
244966_at |
ATCG00600
|
PETG |
-0.257 |
-0.024 |
| 177 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.253 |
-0.408 |
| 178 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.251 |
-0.438 |
| 179 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.245 |
-0.161 |
| 180 |
244965_at |
ATCG00590
|
ORF31 |
-0.244 |
-0.198 |
| 181 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.241 |
0.081 |
| 182 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
-0.240 |
0.266 |
| 183 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.239 |
-0.296 |
| 184 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.238 |
0.434 |
| 185 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.232 |
0.021 |
| 186 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.230 |
0.436 |
| 187 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.226 |
-0.390 |
| 188 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
-0.226 |
0.012 |
| 189 |
257330_at |
ATMG01290
|
ORF111C |
-0.226 |
0.046 |
| 190 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
-0.226 |
-0.169 |
| 191 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.225 |
0.018 |
| 192 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
-0.225 |
-0.069 |
| 193 |
245000_at |
ATCG00210
|
YCF6 |
-0.219 |
-0.022 |
| 194 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
-0.217 |
-0.140 |
| 195 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.215 |
0.106 |
| 196 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.214 |
0.027 |
| 197 |
259723_at |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
-0.210 |
0.067 |
| 198 |
256926_at |
AT3G22540
|
unknown |
-0.209 |
-0.171 |
| 199 |
262711_at |
AT1G16500
|
unknown |
-0.208 |
0.009 |
| 200 |
244932_at |
ATCG01060
|
PSAC |
-0.205 |
-0.060 |
| 201 |
255825_at |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
-0.205 |
-0.154 |
| 202 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
-0.203 |
-0.163 |
| 203 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
-0.203 |
-0.130 |
| 204 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.201 |
-0.171 |
| 205 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
-0.199 |
0.051 |
| 206 |
254022_at |
AT4G25750
|
ABCG4, ATP-binding cassette G4 |
-0.197 |
0.027 |
| 207 |
256066_at |
AT1G06980
|
unknown |
-0.195 |
-0.105 |
| 208 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.190 |
-0.069 |
| 209 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.190 |
-0.120 |
| 210 |
244934_at |
ATCG01080
|
NDHG |
-0.185 |
-0.034 |
| 211 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.183 |
-0.294 |
| 212 |
251067_at |
AT5G01910
|
unknown |
-0.182 |
-0.232 |
| 213 |
257456_at |
AT2G18120
|
SRS4, SHI-related sequence 4 |
-0.179 |
-0.162 |
| 214 |
254145_at |
AT4G24700
|
unknown |
-0.178 |
-0.303 |
| 215 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.178 |
-0.095 |
| 216 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.175 |
0.355 |
| 217 |
259857_at |
AT1G68430
|
unknown |
-0.174 |
-0.057 |
| 218 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.173 |
-0.094 |
| 219 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.172 |
-0.188 |
| 220 |
244972_at |
ATCG00680
|
PSBB, photosystem II reaction center protein B |
-0.171 |
-0.159 |
| 221 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.170 |
0.051 |
| 222 |
263769_at |
AT2G06390
|
transposable element gene |
-0.170 |
0.001 |
| 223 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
-0.169 |
-0.060 |
| 224 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
-0.168 |
-0.087 |
| 225 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
-0.168 |
-0.013 |
| 226 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.167 |
-0.255 |
| 227 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.167 |
-0.163 |
| 228 |
262811_at |
AT1G11700
|
unknown |
-0.167 |
0.190 |
| 229 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.165 |
-0.209 |
| 230 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.165 |
-0.080 |
| 231 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
-0.163 |
-0.039 |
| 232 |
245019_at |
ATCG00530
|
YCF10 |
-0.162 |
-0.071 |
| 233 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
-0.161 |
-0.200 |
| 234 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.160 |
-0.238 |
| 235 |
254574_at |
AT4G19430
|
unknown |
-0.160 |
0.065 |
| 236 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.159 |
-0.024 |
| 237 |
245401_at |
AT4G17670
|
unknown |
-0.159 |
0.145 |
| 238 |
248213_at |
AT5G53660
|
AtGRF7, growth-regulating factor 7, GRF7, growth-regulating factor 7 |
-0.158 |
-0.048 |
| 239 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.158 |
-0.451 |
| 240 |
262388_at |
AT1G49320
|
unknown |
-0.156 |
-0.258 |
| 241 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
-0.156 |
-0.098 |
| 242 |
251388_at |
AT3G60840
|
MAP65-4, microtubule-associated protein 65-4 |
-0.156 |
-0.172 |
| 243 |
244995_at |
ATCG00150
|
ATPI |
-0.155 |
-0.072 |
| 244 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.155 |
-0.154 |
| 245 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
-0.154 |
-0.048 |
| 246 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.153 |
-0.137 |
| 247 |
245075_at |
AT2G23180
|
CYP96A1, cytochrome P450, family 96, subfamily A, polypeptide 1 |
-0.153 |
0.008 |
| 248 |
261588_at |
AT1G01670
|
[RING/U-box superfamily protein] |
-0.153 |
0.006 |
| 249 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.152 |
-0.274 |
| 250 |
260081_at |
AT1G78170
|
unknown |
-0.151 |
-0.246 |
| 251 |
254096_at |
AT4G25150
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.151 |
-0.230 |
| 252 |
252010_at |
AT3G52740
|
unknown |
-0.151 |
-0.232 |
| 253 |
267639_at |
AT2G42200
|
AtSPL9, SPL9, squamosa promoter binding protein-like 9 |
-0.150 |
-0.294 |
| 254 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.150 |
0.118 |
| 255 |
247878_at |
AT5G57760
|
unknown |
-0.149 |
-0.074 |
| 256 |
261221_at |
AT1G19960
|
unknown |
-0.148 |
0.217 |
| 257 |
256538_x_at |
AT3G32900
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G47260.1)] |
-0.147 |
-0.003 |
| 258 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.146 |
-0.092 |
| 259 |
260483_at |
AT1G11000
|
ATMLO4, MILDEW RESISTANCE LOCUS O 4, MLO4, MILDEW RESISTANCE LOCUS O 4 |
-0.146 |
0.045 |
| 260 |
267012_at |
AT2G39220
|
PLA IIB, PLP6, PATATIN-like protein 6 |
-0.145 |
-0.129 |
| 261 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
-0.145 |
0.022 |
| 262 |
260259_at |
AT1G74300
|
[alpha/beta-Hydrolases superfamily protein] |
-0.143 |
0.032 |
| 263 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
-0.143 |
0.004 |
| 264 |
251644_at |
AT3G57540
|
[Remorin family protein] |
-0.142 |
0.028 |
| 265 |
257925_at |
AT3G23170
|
unknown |
-0.141 |
0.190 |
| 266 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.141 |
-0.013 |
| 267 |
260558_at |
AT2G43600
|
[Chitinase family protein] |
-0.141 |
-0.147 |
| 268 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.141 |
-0.083 |
| 269 |
248333_at |
AT5G52390
|
[PAR1 protein] |
-0.140 |
0.348 |
| 270 |
251289_at |
AT3G61830
|
ARF18, auxin response factor 18 |
-0.140 |
-0.041 |
| 271 |
251363_at |
AT3G61250
|
AtMYB17, myb domain protein 17, LMI2, LATE MERISTEM IDENTITY2, MYB17, myb domain protein 17 |
-0.139 |
-0.161 |
| 272 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.139 |
-0.146 |
| 273 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
-0.138 |
-0.277 |
| 274 |
262558_at |
AT1G31335
|
unknown |
-0.138 |
-0.204 |
| 275 |
258082_at |
AT3G25905
|
CLE27, CLAVATA3/ESR-RELATED 27 |
-0.138 |
-0.050 |
| 276 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.138 |
-0.052 |
| 277 |
260075_at |
AT1G73700
|
[MATE efflux family protein] |
-0.137 |
-0.155 |
| 278 |
248674_at |
AT5G48800
|
[Phototropic-responsive NPH3 family protein] |
-0.137 |
-0.134 |
| 279 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.136 |
-0.213 |
| 280 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
-0.135 |
0.069 |
| 281 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.134 |
-0.202 |
| 282 |
245990_at |
AT5G20640
|
unknown |
-0.134 |
-0.160 |
| 283 |
246997_at |
AT5G67390
|
unknown |
-0.134 |
-0.347 |
| 284 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-0.134 |
-0.211 |
| 285 |
245785_at |
AT1G32180
|
ATCSLD6, cellulose synthase-like D6, CSLD6, CELLULOSE SYNTHASE LIKE D6, CSLD6, cellulose synthase-like D6 |
-0.133 |
0.024 |
| 286 |
262590_at |
AT1G15100
|
RHA2A, RING-H2 finger A2A |
-0.133 |
-0.041 |
| 287 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.133 |
-0.154 |
| 288 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.132 |
-0.187 |
| 289 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.131 |
0.014 |
| 290 |
256587_at |
AT3G28780
|
unknown |
-0.130 |
-0.041 |
| 291 |
248028_at |
AT5G55620
|
unknown |
-0.130 |
-0.235 |
| 292 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.130 |
-0.086 |
| 293 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
-0.130 |
0.016 |
| 294 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.130 |
-0.319 |
| 295 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
-0.129 |
-0.143 |
| 296 |
259970_at |
AT1G76570
|
AtLHCB7, LHCB7, light-harvesting complex B7 |
-0.129 |
-0.090 |
| 297 |
254632_at |
AT4G18630
|
unknown |
-0.129 |
-0.251 |
| 298 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
-0.129 |
-0.071 |
| 299 |
264590_at |
AT2G17710
|
unknown |
-0.128 |
0.082 |
| 300 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.128 |
0.074 |
| 301 |
249555_at |
AT5G38300
|
unknown |
-0.128 |
-0.158 |
| 302 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.128 |
-0.005 |
| 303 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.127 |
-0.424 |
| 304 |
254319_at |
AT4G22560
|
unknown |
-0.126 |
-0.078 |
| 305 |
255702_at |
AT4G00230
|
XSP1, xylem serine peptidase 1 |
-0.126 |
-0.099 |
| 306 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
-0.125 |
-0.036 |
| 307 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.125 |
0.317 |
| 308 |
248097_at |
AT5G55260
|
PPX-2, PROTEIN PHOSPHATASE X -2, PPX2, protein phosphatase X 2 |
-0.124 |
-0.105 |
| 309 |
253883_at |
AT4G27660
|
unknown |
-0.124 |
-0.087 |
| 310 |
265831_at |
AT2G14460
|
unknown |
-0.124 |
-0.148 |
| 311 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.123 |
-0.036 |
| 312 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.123 |
-0.158 |
| 313 |
266307_at |
AT2G27000
|
CYP705A8, cytochrome P450, family 705, subfamily A, polypeptide 8 |
-0.123 |
-0.113 |
| 314 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
-0.122 |
-0.091 |
| 315 |
259978_at |
AT1G76540
|
CDKB2;1, cyclin-dependent kinase B2;1 |
-0.122 |
-0.195 |
| 316 |
257916_at |
AT3G23210
|
bHLH34, basic Helix-Loop-Helix 34 |
-0.122 |
-0.135 |
| 317 |
245197_at |
AT1G67800
|
[Copine (Calcium-dependent phospholipid-binding protein) family] |
-0.122 |
0.181 |