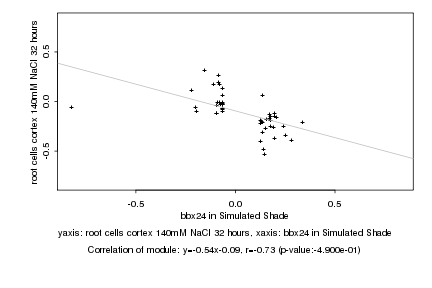
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
bbx24 in Simulated Shade |
Log2 signal ratio
root cells cortex 140mM NaCl 32 hours |
| 1 |
254534_at |
AT4G19680
|
ATIRT2, IRON REGULATED TRANSPORTER 2, IRT2, iron regulated transporter 2 |
0.335 |
-0.206 |
| 2 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
0.279 |
-0.385 |
| 3 |
259064_at |
AT3G07490
|
AGD11, ARF-GAP domain 11, AtCML3, CML3, calmodulin-like 3 |
0.251 |
-0.336 |
| 4 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
0.239 |
-0.242 |
| 5 |
250778_at |
AT5G05500
|
MOP10 |
0.202 |
-0.157 |
| 6 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.195 |
-0.142 |
| 7 |
259964_at |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
0.194 |
-0.369 |
| 8 |
258362_at |
AT3G14280
|
unknown |
0.194 |
-0.113 |
| 9 |
253931_at |
AT4G26770
|
[Phosphatidate cytidylyltransferase family protein] |
0.191 |
-0.257 |
| 10 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.175 |
-0.145 |
| 11 |
258178_at |
AT3G21680
|
unknown |
0.174 |
-0.182 |
| 12 |
263614_at |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
0.172 |
-0.248 |
| 13 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.170 |
-0.165 |
| 14 |
252187_at |
AT3G50850
|
[Putative methyltransferase family protein] |
0.169 |
-0.127 |
| 15 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.151 |
-0.177 |
| 16 |
256913_at |
AT3G23870
|
unknown |
0.149 |
-0.264 |
| 17 |
254467_at |
AT4G20450
|
[Leucine-rich repeat protein kinase family protein] |
0.144 |
-0.531 |
| 18 |
254173_at |
AT4G24580
|
REN1, ROP1 ENHANCER 1 |
0.136 |
-0.479 |
| 19 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
0.133 |
-0.211 |
| 20 |
247581_at |
AT5G61350
|
[Protein kinase superfamily protein] |
0.131 |
-0.307 |
| 21 |
260558_at |
AT2G43600
|
[Chitinase family protein] |
0.131 |
0.069 |
| 22 |
259291_at |
AT3G11550
|
CASP2, Casparian strip membrane domain protein 2 |
0.130 |
-0.192 |
| 23 |
263546_at |
AT2G21550
|
[Bifunctional dihydrofolate reductase/thymidylate synthase] |
0.122 |
-0.221 |
| 24 |
251107_at |
AT5G01610
|
unknown |
0.121 |
-0.189 |
| 25 |
261647_at |
AT1G27740
|
RSL4, root hair defective 6-like 4 |
0.121 |
-0.395 |
| 26 |
260956_at |
AT1G06040
|
BBX24, B-box domain protein 24, STO, SALT TOLERANCE |
-0.827 |
-0.056 |
| 27 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.222 |
0.121 |
| 28 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.202 |
-0.060 |
| 29 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.199 |
-0.097 |
| 30 |
251005_at |
AT5G02590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.156 |
0.318 |
| 31 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.111 |
0.177 |
| 32 |
250174_at |
AT5G14380
|
AGP6, arabinogalactan protein 6 |
-0.100 |
-0.035 |
| 33 |
261936_at |
AT1G22600
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.098 |
-0.115 |
| 34 |
266677_at |
AT2G29820
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.096 |
-0.117 |
| 35 |
255034_at |
AT4G09540
|
[copia-like retrotransposon family, has a 1.7e-43 P-value blast match to GB:AAC64917 gag-pol polyprotein (Ty1_Copia-element) (Glycine max)] |
-0.091 |
-0.008 |
| 36 |
258318_at |
AT3G22680
|
RDM1, RNA-DIRECTED DNA METHYLATION 1 |
-0.087 |
0.198 |
| 37 |
246274_at |
AT4G36620
|
GATA19, GATA transcription factor 19, HANL2, hanaba taranu like 2 |
-0.086 |
0.264 |
| 38 |
250014_at |
AT5G17990
|
pat1, PHOSPHORIBOSYLANTHRANILATE TRANSFERASE 1, TRP1, tryptophan biosynthesis 1 |
-0.083 |
-0.000 |
| 39 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.083 |
0.180 |
| 40 |
246897_at |
AT5G25560
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.080 |
-0.025 |
| 41 |
253869_at |
AT4G27510
|
unknown |
-0.069 |
-0.063 |
| 42 |
258996_at |
AT3G01800
|
[Ribosome recycling factor] |
-0.069 |
0.140 |
| 43 |
261737_at |
AT1G47885
|
[Ribonuclease inhibitor] |
-0.069 |
-0.077 |
| 44 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.068 |
-0.004 |
| 45 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
-0.068 |
-0.023 |
| 46 |
253544_at |
AT4G31280
|
unknown |
-0.068 |
-0.010 |
| 47 |
264737_at |
AT1G62210
|
unknown |
-0.066 |
-0.065 |
| 48 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
-0.065 |
0.062 |
| 49 |
260021_at |
AT1G30010
|
nMAT1, nuclear maturase 1 |
-0.065 |
-0.028 |
| 50 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.065 |
-0.097 |