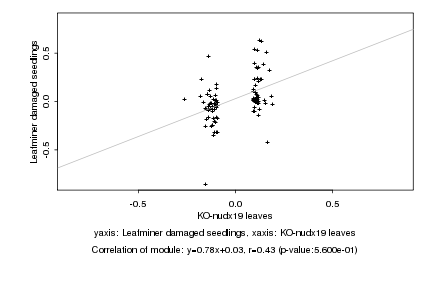
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
KO-nudx19 leaves |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.186 |
-0.023 |
| 2 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.181 |
0.062 |
| 3 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.175 |
0.329 |
| 4 |
248186_at |
AT5G53880
|
unknown |
0.163 |
-0.423 |
| 5 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.156 |
0.514 |
| 6 |
249287_at |
AT5G41250
|
[Exostosin family protein] |
0.152 |
-0.013 |
| 7 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.145 |
0.014 |
| 8 |
250028_at |
AT5G18130
|
unknown |
0.141 |
0.388 |
| 9 |
267209_at |
AT2G30930
|
unknown |
0.134 |
0.235 |
| 10 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.134 |
0.621 |
| 11 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.125 |
0.228 |
| 12 |
260420_at |
AT1G69610
|
[FUNCTIONS IN: structural constituent of ribosome] |
0.120 |
0.633 |
| 13 |
250951_at |
AT5G03550
|
unknown |
0.119 |
-0.079 |
| 14 |
246929_at |
AT5G25210
|
unknown |
0.118 |
0.213 |
| 15 |
251165_at |
AT3G63340
|
[Protein phosphatase 2C family protein] |
0.117 |
-0.139 |
| 16 |
255335_at |
AT4G04430
|
[CACTA-like transposase family (Ptta/En/Spm), has a 8.6e-122 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.116 |
-0.018 |
| 17 |
260139_at |
AT1G66380
|
AtMYB114, myb domain protein 114, MYB114, myb domain protein 114 |
0.115 |
0.044 |
| 18 |
257917_at |
AT3G23220
|
ESE1, ethylene and salt inducible 1 |
0.115 |
0.354 |
| 19 |
248569_at |
AT5G49770
|
[Leucine-rich repeat protein kinase family protein] |
0.114 |
0.003 |
| 20 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
0.113 |
0.535 |
| 21 |
264447_at |
AT1G27300
|
unknown |
0.112 |
0.241 |
| 22 |
255171_at |
AT4G07990
|
[Chaperone DnaJ-domain superfamily protein] |
0.112 |
0.069 |
| 23 |
264851_at |
AT2G17290
|
ATCDPK3, ARABIDOPSIS THALIANA CALMODULIN-DOMAIN PROTEIN KINASE 3, ATCPK6, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 6, CPK6, calcium dependent protein kinase 6 |
0.112 |
0.347 |
| 24 |
251606_at |
AT3G57840
|
[Plant self-incompatibility protein S1 family] |
0.110 |
-0.001 |
| 25 |
266787_at |
AT2G28990
|
[Leucine-rich repeat protein kinase family protein] |
0.110 |
-0.020 |
| 26 |
246941_at |
AT5G25415
|
unknown |
0.109 |
0.027 |
| 27 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.108 |
0.354 |
| 28 |
251247_at |
AT3G62140
|
unknown |
0.107 |
0.049 |
| 29 |
263950_at |
AT2G36020
|
HVA22J, HVA22-like protein J |
0.106 |
-0.004 |
| 30 |
249308_at |
AT5G41350
|
[RING/U-box superfamily protein] |
0.105 |
0.079 |
| 31 |
264283_at |
AT1G61850
|
[phospholipases] |
0.103 |
0.019 |
| 32 |
257471_at |
AT1G59530
|
ATBZIP4, basic leucine-zipper 4, bZIP4, basic leucine-zipper 4 |
0.101 |
0.025 |
| 33 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
0.101 |
-0.001 |
| 34 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
0.101 |
0.173 |
| 35 |
265874_at |
AT2G01640
|
unknown |
0.101 |
0.101 |
| 36 |
262268_at |
AT1G42410
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.0e-124 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.099 |
0.103 |
| 37 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.096 |
0.236 |
| 38 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
0.096 |
0.005 |
| 39 |
251908_at |
AT3G53770
|
[late embryogenesis abundant 3 (LEA3) family protein] |
0.095 |
0.023 |
| 40 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.095 |
0.229 |
| 41 |
260600_at |
AT1G55950
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
0.095 |
0.009 |
| 42 |
246413_at |
AT1G77310
|
unknown |
0.095 |
-0.103 |
| 43 |
267345_at |
AT2G44240
|
unknown |
0.094 |
-0.061 |
| 44 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.094 |
0.399 |
| 45 |
256586_at |
AT3G28770
|
unknown |
0.093 |
0.015 |
| 46 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.093 |
0.548 |
| 47 |
249586_at |
AT5G37840
|
unknown |
0.092 |
0.133 |
| 48 |
265718_at |
AT2G03340
|
WRKY3, WRKY DNA-binding protein 3 |
0.092 |
-0.101 |
| 49 |
262179_at |
AT1G77980
|
AGL66, AGAMOUS-like 66 |
0.090 |
0.038 |
| 50 |
245556_at |
AT4G15400
|
ABS1, ABNORMAL SHOOT 1, BIA1, BRASSINOSTEROID INACTIVATOR1 |
0.090 |
0.014 |
| 51 |
246761_at |
AT5G27980
|
[Seed maturation protein] |
0.089 |
0.017 |
| 52 |
259110_at |
AT3G05570
|
unknown |
0.089 |
0.107 |
| 53 |
255463_at |
AT4G02960
|
ATRE2, retro element 2, RE2, retro element 2 |
0.089 |
0.028 |
| 54 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.264 |
0.024 |
| 55 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.184 |
0.059 |
| 56 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.176 |
0.231 |
| 57 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.167 |
-0.004 |
| 58 |
254574_at |
AT4G19430
|
unknown |
-0.157 |
-0.848 |
| 59 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
-0.155 |
-0.066 |
| 60 |
253723_at |
AT4G29240
|
[Leucine-rich repeat (LRR) family protein] |
-0.155 |
-0.256 |
| 61 |
263448_at |
AT2G31660
|
EMA1, enhanced miRNA activity 1, SAD2, SUPER SENSITIVE TO ABA AND DROUGHT2, URM9, UNARMED 9 |
-0.153 |
-0.178 |
| 62 |
250176_at |
AT5G14400
|
CYP724A1, cytochrome P450, family 724, subfamily A, polypeptide 1 |
-0.152 |
-0.078 |
| 63 |
247200_at |
AT5G65120
|
unknown |
-0.147 |
0.082 |
| 64 |
251909_at |
AT3G53790
|
TRFL4, TRF-like 4 |
-0.146 |
0.080 |
| 65 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
-0.143 |
-0.043 |
| 66 |
267159_at |
AT2G37650
|
[GRAS family transcription factor] |
-0.142 |
-0.162 |
| 67 |
257619_at |
AT3G24810
|
ICK3, KRP5, kip-related protein 5 |
-0.142 |
-0.091 |
| 68 |
265865_at |
AT2G01740
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.141 |
-0.078 |
| 69 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.139 |
0.473 |
| 70 |
257931_at |
AT3G17030
|
[Nucleic acid-binding proteins superfamily] |
-0.137 |
-0.024 |
| 71 |
248020_at |
AT5G56490
|
AtGulLO4, GulLO4, L -gulono-1,4-lactone ( L -GulL) oxidase 4 |
-0.137 |
0.114 |
| 72 |
267371_at |
AT2G44510
|
[CDK inhibitor P21 binding protein] |
-0.133 |
-0.020 |
| 73 |
249707_at |
AT5G35640
|
[Putative endonuclease or glycosyl hydrolase] |
-0.131 |
-0.071 |
| 74 |
244923_s_at |
ATMG01020
|
ORF153B |
-0.129 |
0.060 |
| 75 |
251900_at |
AT3G54430
|
SRS6, SHI-related sequence 6 |
-0.128 |
-0.257 |
| 76 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.126 |
-0.012 |
| 77 |
248638_at |
AT5G49070
|
KCS21, 3-ketoacyl-CoA synthase 21 |
-0.123 |
-0.048 |
| 78 |
245569_at |
AT4G14660
|
NRPE7 |
-0.122 |
-0.077 |
| 79 |
260523_at |
AT2G41720
|
EMB2654, EMBRYO DEFECTIVE 2654 |
-0.120 |
-0.238 |
| 80 |
246274_at |
AT4G36620
|
GATA19, GATA transcription factor 19, HANL2, hanaba taranu like 2 |
-0.119 |
-0.101 |
| 81 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.119 |
-0.246 |
| 82 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.117 |
-0.175 |
| 83 |
259823_at |
AT1G66250
|
[O-Glycosyl hydrolases family 17 protein] |
-0.114 |
-0.342 |
| 84 |
259930_at |
AT1G34355
|
ATPS1, PARALLEL SPINDLE 1, PS1, PARALLEL SPINDLE 1 |
-0.114 |
0.027 |
| 85 |
251119_at |
AT3G63510
|
[FMN-linked oxidoreductases superfamily protein] |
-0.112 |
-0.198 |
| 86 |
247344_at |
AT5G63750
|
ARI13, ARIADNE 13, ATARI13, ARABIDOPSIS ARIADNE 13 |
-0.111 |
-0.014 |
| 87 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-0.110 |
-0.317 |
| 88 |
246266_at |
AT1G31817
|
NFD3, NUCLEAR FUSION DEFECTIVE 3 |
-0.109 |
-0.077 |
| 89 |
262596_at |
AT1G15080
|
ATLPP2, LIPID PHOSPHATE PHOSPHATASE 2, ATPAP2, PHOSPHATIDIC ACID PHOSPHATASE 2, LPP2, lipid phosphate phosphatase 2 |
-0.108 |
-0.077 |
| 90 |
262922_at |
AT1G79420
|
unknown |
-0.108 |
-0.043 |
| 91 |
264634_at |
AT1G65670
|
CYP702A1, cytochrome P450, family 702, subfamily A, polypeptide 1 |
-0.107 |
0.065 |
| 92 |
254986_at |
AT4G10640
|
IQD16, IQ-domain 16 |
-0.103 |
0.018 |
| 93 |
263146_at |
AT1G53940
|
GLIP2, GDSL-motif lipase 2 |
-0.103 |
-0.023 |
| 94 |
257744_at |
AT3G29230
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.103 |
-0.209 |
| 95 |
253589_at |
AT4G30825
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.102 |
-0.317 |
| 96 |
260392_at |
AT1G74030
|
ENO1, enolase 1 |
-0.101 |
0.179 |
| 97 |
253017_at |
AT4G37970
|
ATCAD6, CAD6, cinnamyl alcohol dehydrogenase 6 |
-0.100 |
0.143 |
| 98 |
251631_at |
AT3G57430
|
OTP84, ORGANELLE TRANSCRIPT PROCESSING 84 |
-0.100 |
-0.057 |
| 99 |
249182_at |
AT5G42960
|
unknown |
-0.100 |
-0.035 |
| 100 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
-0.098 |
0.015 |
| 101 |
247628_at |
AT5G60400
|
unknown |
-0.098 |
-0.161 |
| 102 |
245937_at |
AT5G19750
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.097 |
-0.037 |
| 103 |
267304_at |
AT2G30080
|
ATZIP6, ZIP6 |
-0.096 |
-0.174 |
| 104 |
257543_at |
AT3G28960
|
[Transmembrane amino acid transporter family protein] |
-0.095 |
-0.317 |
| 105 |
265195_at |
AT2G36730
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.095 |
-0.005 |