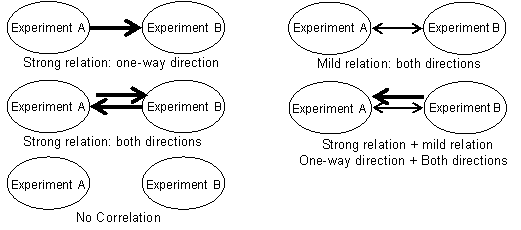
3.1:Method
Microarray data is noisy "snapshot" of transcriptional state of cells. Detecting biological correlations among gene expression profiles from multiple laboratories on a large scale remains difficult.
Here, we applied a module (sets of genes working in the same biological pathway)-based correlation analysis in combination with a network analysis to Arabidopsis data and developed a "relation map", which represents relationships among DNA microarray experiments on a large scale. In each experiment, the gene expression responses of modules are closely correlated with the status of specific biological actions. Therefore, one can assume that samples sharing a common response in a module also share a related biological action or response. According to following idea, we chose modules from each experiment and calculated correlations between gene expression profiles by using it.
A. Dataset structure
Table 1 Example of data matrix of microarray data.
| Experiment A | Experiment B | Experiment C | ||||
| gene1 | a** | a* | a | |||
| gene2 | b | a | c | |||
| gene3 | c | c | f | |||
| gene4 | d | b | d | |||
*Characters (a, b, c, d..) in 'Table 1' indicate signal ratio of each gene, namely, log2 (average of signal intensity on treatment / average of signal intensity on control)
**Bold characters indicate genes contained in the module.
For a module, induced or repressed genes in each experiment were selected by statistical methods based on the P value of Student's t-test and the fold-change in induction levels between treatment and control samples. The thresholds for selecting modules were set as follows: detection P value for AG was 0.1 (detected in MAS5 method), P value of Student's t-test for AG was 0.05 (significantly induced or repressed), detection P value for ATH1 was 0.01, P value of Student's t-test for ATH1 was 0.01, P value of Student's t-test for Aragene11ST was 0.01. The size of module is always more than 50. If it is less than 50, then AtCAST selects less significant probes. These genes represent the biological status of the samples analyzed in each experiment.
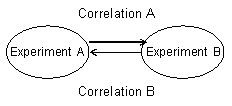
B. Calculation of correlation between experiments
Spearman’s rank-order correlation coefficients (SCCs) to estimate relationships in gene expression profiles between experiments based on modules with bold characters.
Correlation A: From Experiment A to Experiment B
Experiment A (a, b, c) vs. Experiment B (a, a, c)
(Using a module of Experiment A)
Correlation B: From Experiment A to Experiment B
Experiment B (a, b) vs. Experiment A (b, d)
(Using a module of Experiment B)
C: Drawing relation maps
Combining results based on SCC.