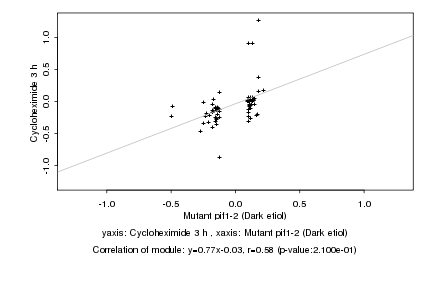
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant pif1-2 (Dark etiol) |
Log2 signal ratio
Cycloheximide 3 h |
| 1 |
260742_at |
AT1G15050
|
IAA34, indole-3-acetic acid inducible 34 |
0.214 |
0.184 |
| 2 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.175 |
1.280 |
| 3 |
246773_at |
AT5G27510
|
[Protein kinase superfamily protein] |
0.174 |
0.168 |
| 4 |
258541_at |
AT3G07000
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.173 |
0.390 |
| 5 |
251392_at |
AT3G60880
|
DHDPS, DIHYDRODIPICOLINATE SYNTHASE, DHDPS1, dihydrodipicolinate synthase 1 |
0.165 |
-0.198 |
| 6 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
0.159 |
-0.203 |
| 7 |
255285_at |
AT4G04630
|
unknown |
0.148 |
-0.033 |
| 8 |
266361_at |
AT2G32450
|
[Calcium-binding tetratricopeptide family protein] |
0.143 |
0.050 |
| 9 |
253261_at |
AT4G34430
|
ATSWI3D, SWITCH/SUCROSE NONFERMENTING 3D, CHB3 |
0.134 |
0.030 |
| 10 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.133 |
0.023 |
| 11 |
263189_at |
AT1G36100
|
[myosin heavy chain-related] |
0.133 |
0.032 |
| 12 |
245186_at |
AT1G67710
|
ARR11, response regulator 11 |
0.132 |
0.067 |
| 13 |
246639_x_at |
AT5G34895
|
[similar to heat shock protein binding [Arabidopsis thaliana] (TAIR:AT1G32830.1)] |
0.129 |
0.014 |
| 14 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
0.127 |
0.913 |
| 15 |
254421_at |
AT4G21550
|
VAL3, VP1/ABI3-like 3 |
0.121 |
-0.033 |
| 16 |
263290_at |
AT2G10930
|
unknown |
0.116 |
0.028 |
| 17 |
248240_at |
AT5G53950
|
ANAC098, Arabidopsis NAC domain containing protein 98, ATCUC2, CUP-SHAPED COTYLEDON 2, CUC2, CUP-SHAPED COTYLEDON 2 |
0.112 |
-0.056 |
| 18 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.112 |
-0.259 |
| 19 |
265585_at |
AT2G20000
|
CDC27b, HBT, HOBBIT |
0.111 |
-0.094 |
| 20 |
250938_at |
AT5G03180
|
[RING/U-box superfamily protein] |
0.110 |
0.045 |
| 21 |
264379_at |
AT2G25200
|
unknown |
0.110 |
0.063 |
| 22 |
251320_at |
AT3G61530
|
PANB2 |
0.109 |
-0.066 |
| 23 |
257453_at |
AT1G65130
|
[Ubiquitin carboxyl-terminal hydrolase-related protein] |
0.105 |
-0.031 |
| 24 |
264345_at |
AT1G11915
|
unknown |
0.104 |
0.015 |
| 25 |
267487_at |
AT2G19100
|
[non-LTR retrotransposon family (LINE), has a 3.2e-33 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
0.103 |
-0.062 |
| 26 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.101 |
-0.306 |
| 27 |
267620_at |
AT2G39640
|
[glycosyl hydrolase family 17 protein] |
0.098 |
0.069 |
| 28 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.098 |
-0.114 |
| 29 |
256006_at |
AT1G34070
|
unknown |
0.095 |
0.010 |
| 30 |
259166_at |
AT3G01670
|
AtSEOR2, Arabidopsis thaliana sieve element occlusion-related 2, SEOa, sieve element occlusion a, SEOR2, sieve element occlusion-related 2 |
0.094 |
-0.162 |
| 31 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
0.094 |
-0.233 |
| 32 |
245447_at |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
0.094 |
0.913 |
| 33 |
246703_at |
AT5G28080
|
WNK9 |
0.091 |
0.007 |
| 34 |
247876_at |
AT5G57730
|
unknown |
0.091 |
0.021 |
| 35 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
-0.504 |
-0.227 |
| 36 |
255805_at |
AT4G10240
|
bbx23, B-box domain protein 23 |
-0.497 |
-0.066 |
| 37 |
247878_at |
AT5G57760
|
unknown |
-0.275 |
-0.458 |
| 38 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.254 |
-0.331 |
| 39 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
-0.249 |
-0.010 |
| 40 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
-0.234 |
-0.228 |
| 41 |
253233_at |
AT4G34290
|
[SWIB/MDM2 domain superfamily protein] |
-0.232 |
-0.182 |
| 42 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.217 |
-0.312 |
| 43 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.208 |
-0.204 |
| 44 |
259529_at |
AT1G12400
|
[Nucleotide excision repair, TFIIH, subunit TTDA] |
-0.185 |
-0.398 |
| 45 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.183 |
-0.168 |
| 46 |
245354_at |
AT4G17600
|
LIL3:1 |
-0.182 |
-0.135 |
| 47 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
-0.179 |
-0.043 |
| 48 |
249830_at |
AT5G23300
|
PYRD, pyrimidine d |
-0.175 |
-0.143 |
| 49 |
249010_at |
AT5G44580
|
unknown |
-0.172 |
0.041 |
| 50 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.161 |
-0.309 |
| 51 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.160 |
-0.238 |
| 52 |
255243_at |
AT4G05590
|
unknown |
-0.158 |
-0.083 |
| 53 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
-0.156 |
-0.100 |
| 54 |
244966_at |
ATCG00600
|
PETG |
-0.153 |
-0.258 |
| 55 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
-0.150 |
-0.105 |
| 56 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.149 |
-0.344 |
| 57 |
251958_at |
AT3G53560
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.148 |
-0.138 |
| 58 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.148 |
-0.277 |
| 59 |
254228_at |
AT4G23620
|
[Ribosomal protein L25/Gln-tRNA synthetase, anti-codon-binding domain] |
-0.143 |
-0.087 |
| 60 |
266608_at |
AT2G35500
|
SKL2, shikimate kinase-like 2 |
-0.142 |
-0.107 |
| 61 |
266587_at |
AT2G14880
|
[SWIB/MDM2 domain superfamily protein] |
-0.141 |
-0.202 |
| 62 |
249826_at |
AT5G23310
|
FSD3, Fe superoxide dismutase 3 |
-0.134 |
-0.100 |
| 63 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
-0.130 |
0.151 |
| 64 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.130 |
-0.141 |
| 65 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.129 |
-0.120 |
| 66 |
265704_at |
AT2G03420
|
unknown |
-0.129 |
-0.237 |
| 67 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.127 |
-0.864 |