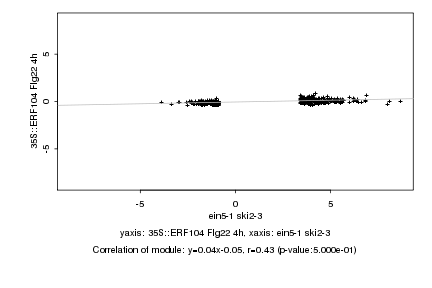
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ein5-1 ski2-3 |
Log2 signal ratio
35S::ERF104 Flg22 4h |
| 1 |
AT3G13960 |
AT3G13960
|
AtGRF5, growth-regulating factor 5, GRF5, growth-regulating factor 5 |
8.658 |
0.039 |
| 2 |
AT5G57130 |
AT5G57130
|
SMXL5, SMAX1-like 5 |
8.095 |
0.026 |
| 3 |
AT4G23750 |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
7.986 |
-0.257 |
| 4 |
AT4G29920 |
AT4G29920
|
SMXL4, SMAX1-like 4 |
7.802 |
no data |
| 5 |
AT3G02840 |
AT3G02840
|
[ARM repeat superfamily protein] |
6.855 |
0.701 |
| 6 |
AT4G34410 |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
6.836 |
0.007 |
| 7 |
AT5G52050 |
AT5G52050
|
[MATE efflux family protein] |
6.824 |
0.179 |
| 8 |
AT4G30280 |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
6.587 |
-0.063 |
| 9 |
AT3G48520 |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
6.450 |
-0.004 |
| 10 |
AT1G65390 |
AT1G65390
|
ATPP2-A5, phloem protein 2 A5, PP2-A5, phloem protein 2 A5 |
6.372 |
0.144 |
| 11 |
AT5G62520 |
AT5G62520
|
SRO5, similar to RCD one 5 |
6.358 |
0.082 |
| 12 |
AT1G12200 |
AT1G12200
|
FMO, flavin monooxygenase |
6.246 |
0.115 |
| 13 |
AT3G16860 |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
6.211 |
0.222 |
| 14 |
AT3G23250 |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
6.207 |
0.422 |
| 15 |
AT1G76650 |
AT1G76650
|
CML38, calmodulin-like 38 |
6.203 |
0.025 |
| 16 |
AT5G01100 |
AT5G01100
|
FRB1, FRIABLE 1 |
6.144 |
no data |
| 17 |
AT4G23387 |
AT4G23387
|
MIR845A, microRNA845A |
6.016 |
no data |
| 18 |
AT3G01830 |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
5.996 |
0.468 |
| 19 |
AT5G03210 |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
5.967 |
-0.051 |
| 20 |
AT3G27250 |
AT3G27250
|
unknown |
5.913 |
no data |
| 21 |
AT2G39870 |
AT2G39870
|
unknown |
5.657 |
0.155 |
| 22 |
AT1G01560 |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
5.629 |
0.112 |
| 23 |
AT1G28370 |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
5.623 |
0.129 |
| 24 |
AT1G61340 |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
5.616 |
-0.039 |
| 25 |
AT5G42380 |
AT5G42380
|
CML37, calmodulin like 37 |
5.615 |
0.215 |
| 26 |
AT1G72520 |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
5.596 |
0.190 |
| 27 |
AT4G31800 |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
5.574 |
0.022 |
| 28 |
AT1G02400 |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
5.564 |
0.192 |
| 29 |
AT1G12610 |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
5.520 |
0.038 |
| 30 |
AT2G46630 |
AT2G46630
|
unknown |
5.491 |
-0.027 |
| 31 |
AT2G46400 |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
5.490 |
0.276 |
| 32 |
AT4G01360 |
AT4G01360
|
BPS3, BYPASS 3 |
5.479 |
no data |
| 33 |
AT5G56980 |
AT5G56980
|
unknown |
5.479 |
0.052 |
| 34 |
AT4G27654 |
AT4G27654
|
unknown |
5.474 |
-0.109 |
| 35 |
AT2G27690 |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
5.379 |
0.007 |
| 36 |
AT4G39670 |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
5.352 |
0.421 |
| 37 |
AT4G29780 |
AT4G29780
|
unknown |
5.339 |
0.272 |
| 38 |
AT1G11700 |
AT1G11700
|
unknown |
5.266 |
-0.102 |
| 39 |
AT2G27080 |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
5.263 |
0.168 |
| 40 |
AT5G64870 |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
5.250 |
0.104 |
| 41 |
AT3G08720 |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
5.245 |
0.124 |
| 42 |
AT4G37590 |
AT4G37590
|
MEL1, MAB4/ENP/NPY1-LIKE 1, NPY5, NAKED PINS IN YUC MUTANTS 5 |
5.221 |
-0.005 |
| 43 |
AT2G24600 |
AT2G24600
|
[Ankyrin repeat family protein] |
5.210 |
0.189 |
| 44 |
AT4G36950 |
AT4G36950
|
MAPKKK21, mitogen-activated protein kinase kinase kinase 21 |
5.199 |
no data |
| 45 |
AT2G22880 |
AT2G22880
|
[VQ motif-containing protein] |
5.192 |
0.317 |
| 46 |
AT2G18570 |
AT2G18570
|
[UDP-Glycosyltransferase superfamily protein] |
5.187 |
no data |
| 47 |
AT4G25810 |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
5.177 |
-0.001 |
| 48 |
AT1G73870 |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
5.163 |
-0.066 |
| 49 |
AT2G35710 |
AT2G35710
|
PGSIP7, plant glycogenin-like starch initiation protein 7 |
5.150 |
0.210 |
| 50 |
AT1G19020 |
AT1G19020
|
unknown |
5.139 |
0.161 |
| 51 |
AT1G61800 |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
5.030 |
0.337 |
| 52 |
AT2G44840 |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
5.009 |
0.049 |
| 53 |
AT3G25780 |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
4.980 |
0.055 |
| 54 |
AT2G25460 |
AT2G25460
|
unknown |
4.968 |
0.156 |
| 55 |
AT1G17420 |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
4.930 |
0.221 |
| 56 |
AT3G50930 |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
4.915 |
0.304 |
| 57 |
AT4G14370 |
AT4G14370
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
4.880 |
no data |
| 58 |
AT4G19230 |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
4.877 |
-0.161 |
| 59 |
AT2G32210 |
AT2G32210
|
unknown |
4.874 |
no data |
| 60 |
AT3G46090 |
AT3G46090
|
ZAT7 |
4.848 |
no data |
| 61 |
AT3G10930 |
AT3G10930
|
unknown |
4.837 |
0.563 |
| 62 |
AT3G29000 |
AT3G29000
|
[Calcium-binding EF-hand family protein] |
4.837 |
no data |
| 63 |
AT5G66650 |
AT5G66650
|
unknown |
4.834 |
0.111 |
| 64 |
AT2G34930 |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
4.829 |
-0.032 |
| 65 |
AT5G19470 |
AT5G19470
|
atnudt24, nudix hydrolase homolog 24, NUDT24, nudix hydrolase homolog 24, NUDX24, nudix hydrolase homolog 24 |
4.819 |
no data |
| 66 |
AT5G01830 |
AT5G01830
|
SAUR21, SMALL AUXIN UP RNA 21 |
4.797 |
0.391 |
| 67 |
AT4G37730 |
AT4G37730
|
AtbZIP7, basic leucine-zipper 7, bZIP7, basic leucine-zipper 7 |
4.786 |
-0.098 |
| 68 |
AT1G13360 |
AT1G13360
|
unknown |
4.764 |
no data |
| 69 |
AT1G80840 |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
4.743 |
0.094 |
| 70 |
AT1G16370 |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
4.702 |
0.093 |
| 71 |
AT1G62670 |
AT1G62670
|
RPF2, rna processing factor 2 |
4.700 |
-0.158 |
| 72 |
AT1G19180 |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
4.664 |
0.077 |
| 73 |
AT3G19970 |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
4.649 |
0.222 |
| 74 |
AT1G30135 |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
4.640 |
-0.192 |
| 75 |
AT2G42760 |
AT2G42760
|
unknown |
4.637 |
0.059 |
| 76 |
AT2G35930 |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
4.619 |
0.439 |
| 77 |
AT1G09932 |
AT1G09932
|
[Phosphoglycerate mutase family protein] |
4.618 |
no data |
| 78 |
AT2G34600 |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
4.610 |
-0.163 |
| 79 |
AT5G41750 |
AT5G41750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
4.587 |
no data |
| 80 |
AT4G27260 |
AT4G27260
|
GH3.5, WES1 |
4.585 |
-0.051 |
| 81 |
AT5G54710 |
AT5G54710
|
[Ankyrin repeat family protein] |
4.580 |
no data |
| 82 |
AT1G30330 |
AT1G30330
|
ARF6, auxin response factor 6 |
4.520 |
0.015 |
| 83 |
AT1G17380 |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
4.512 |
-0.101 |
| 84 |
AT1G63720 |
AT1G63720
|
unknown |
4.504 |
0.190 |
| 85 |
AT3G01420 |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
4.504 |
0.069 |
| 86 |
AT1G20070 |
AT1G20070
|
unknown |
4.496 |
-0.066 |
| 87 |
AT5G20230 |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
4.488 |
0.106 |
| 88 |
AT5G54060 |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
4.488 |
-0.009 |
| 89 |
AT1G32900 |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
4.488 |
-0.115 |
| 90 |
AT5G52750 |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
4.476 |
0.362 |
| 91 |
AT5G36925 |
AT5G36925
|
unknown |
4.447 |
no data |
| 92 |
AT5G54510 |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
4.441 |
0.025 |
| 93 |
AT4G17230 |
AT4G17230
|
SCL13, SCARECROW-like 13 |
4.417 |
0.064 |
| 94 |
AT5G52760 |
AT5G52760
|
[Copper transport protein family] |
4.416 |
0.329 |
| 95 |
AT3G61630 |
AT3G61630
|
CRF6, cytokinin response factor 6 |
4.415 |
-0.136 |
| 96 |
AT1G19380 |
AT1G19380
|
unknown |
4.401 |
0.141 |
| 97 |
AT3G47500 |
AT3G47500
|
CDF3, cycling DOF factor 3 |
4.389 |
-0.120 |
| 98 |
AT2G30040 |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
4.372 |
-0.050 |
| 99 |
AT1G28480 |
AT1G28480
|
GRX480, roxy19 |
4.365 |
-0.027 |
| 100 |
AT2G20142 |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
4.360 |
0.352 |
| 101 |
AT5G43420 |
AT5G43420
|
[RING/U-box superfamily protein] |
4.360 |
no data |
| 102 |
AT1G61890 |
AT1G61890
|
[MATE efflux family protein] |
4.351 |
-0.136 |
| 103 |
AT3G08860 |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
4.348 |
-0.009 |
| 104 |
AT2G38470 |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
4.343 |
0.248 |
| 105 |
AT1G74430 |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
4.318 |
-0.256 |
| 106 |
AT1G30720 |
AT1G30720
|
[FAD-binding Berberine family protein] |
4.313 |
no data |
| 107 |
AT2G30020 |
AT2G30020
|
[Protein phosphatase 2C family protein] |
4.294 |
no data |
| 108 |
AT3G25870 |
AT3G25870
|
unknown |
4.287 |
0.018 |
| 109 |
AT1G74450 |
AT1G74450
|
unknown |
4.277 |
0.191 |
| 110 |
AT2G32200 |
AT2G32200
|
unknown |
4.270 |
no data |
| 111 |
AT4G10390 |
AT4G10390
|
[Protein kinase superfamily protein] |
4.264 |
0.050 |
| 112 |
AT1G55920 |
AT1G55920
|
ATSERAT2;1, serine acetyltransferase 2;1, SAT1, SERINE ACETYLTRANSFERASE 1, SAT5, SERINE ACETYLTRANSFERASE 5, SERAT2;1, serine acetyltransferase 2;1 |
4.256 |
-0.023 |
| 113 |
AT3G14440 |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
4.255 |
-0.112 |
| 114 |
AT5G65300 |
AT5G65300
|
unknown |
4.246 |
0.159 |
| 115 |
AT1G58420 |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
4.244 |
0.244 |
| 116 |
AT3G02020 |
AT3G02020
|
AK3, aspartate kinase 3 |
4.231 |
no data |
| 117 |
AT2G44500 |
AT2G44500
|
[O-fucosyltransferase family protein] |
4.228 |
0.145 |
| 118 |
AT5G41080 |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
4.197 |
-0.207 |
| 119 |
AT1G14540 |
AT1G14540
|
PER4, peroxidase 4 |
4.196 |
0.891 |
| 120 |
AT1G05340 |
AT1G05340
|
unknown |
4.196 |
0.042 |
| 121 |
AT5G49520 |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
4.182 |
0.250 |
| 122 |
AT5G66070 |
AT5G66070
|
[RING/U-box superfamily protein] |
4.174 |
0.180 |
| 123 |
AT2G28350 |
AT2G28350
|
ARF10, auxin response factor 10 |
4.161 |
0.030 |
| 124 |
AT4G02330 |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
4.149 |
0.348 |
| 125 |
AT1G77640 |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
4.144 |
-0.211 |
| 126 |
AT1G18300 |
AT1G18300
|
atnudt4, nudix hydrolase homolog 4, NUDT4, nudix hydrolase homolog 4 |
4.130 |
no data |
| 127 |
AT2G41010 |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
4.127 |
0.331 |
| 128 |
AT2G27310 |
AT2G27310
|
[F-box family protein] |
4.126 |
0.213 |
| 129 |
AT2G22470 |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
4.124 |
0.032 |
| 130 |
AT5G09800 |
AT5G09800
|
[ARM repeat superfamily protein] |
4.089 |
0.652 |
| 131 |
AT4G18010 |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
4.080 |
0.093 |
| 132 |
AT5G21960 |
AT5G21960
|
[Integrase-type DNA-binding superfamily protein] |
4.069 |
no data |
| 133 |
AT3G48650 |
AT3G48650
|
[pseudogene] |
4.066 |
0.670 |
| 134 |
AT3G55980 |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
4.055 |
0.320 |
| 135 |
AT1G05675 |
AT1G05675
|
[UDP-Glycosyltransferase superfamily protein] |
4.055 |
no data |
| 136 |
AT3G25250 |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
4.050 |
0.267 |
| 137 |
AT1G35140 |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
4.041 |
-0.258 |
| 138 |
AT4G14365 |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
4.015 |
0.281 |
| 139 |
AT3G47340 |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
4.012 |
-0.407 |
| 140 |
AT4G27652 |
AT4G27652
|
unknown |
4.005 |
0.143 |
| 141 |
AT1G14870 |
AT1G14870
|
AtPCR2, PCR2, PLANT CADMIUM RESISTANCE 2 |
3.998 |
no data |
| 142 |
AT2G14290 |
AT2G14290
|
unknown |
3.992 |
0.041 |
| 143 |
AT2G38790 |
AT2G38790
|
unknown |
3.990 |
0.017 |
| 144 |
AT4G32880 |
AT4G32880
|
ATHB-8, homeobox gene 8, ATHB8, HB-8, homeobox gene 8 |
3.979 |
0.012 |
| 145 |
AT1G66090 |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
3.974 |
0.453 |
| 146 |
AT2G19800 |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
3.964 |
-0.255 |
| 147 |
AT1G05575 |
AT1G05575
|
unknown |
3.959 |
0.417 |
| 148 |
AT5G26920 |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
3.957 |
0.583 |
| 149 |
AT5G57560 |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
3.947 |
-0.212 |
| 150 |
AT2G18690 |
AT2G18690
|
unknown |
3.944 |
0.269 |
| 151 |
AT4G16563 |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
3.932 |
-0.410 |
| 152 |
AT2G32190 |
AT2G32190
|
unknown |
3.931 |
no data |
| 153 |
AT5G45340 |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
3.922 |
0.109 |
| 154 |
AT3G52400 |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
3.916 |
0.370 |
| 155 |
AT2G22850 |
AT2G22850
|
AtbZIP6, basic leucine-zipper 6, bZIP6, basic leucine-zipper 6 |
3.914 |
-0.056 |
| 156 |
AT3G19380 |
AT3G19380
|
PUB25, plant U-box 25 |
3.914 |
-0.035 |
| 157 |
AT4G20860 |
AT4G20860
|
[FAD-binding Berberine family protein] |
3.908 |
0.221 |
| 158 |
AT1G66160 |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
3.894 |
0.380 |
| 159 |
AT1G77760 |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
3.891 |
-0.210 |
| 160 |
AT1G19210 |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
3.886 |
0.069 |
| 161 |
AT4G24110 |
AT4G24110
|
unknown |
3.885 |
0.325 |
| 162 |
AT1G56660 |
AT1G56660
|
unknown |
3.884 |
-0.048 |
| 163 |
AT4G12720 |
AT4G12720
|
AtNUDT7, Arabidopsis thaliana Nudix hydrolase homolog 7, GFG1, GROWTH FACTOR GENE 1, NUDT7 |
3.882 |
0.111 |
| 164 |
AT1G10340 |
AT1G10340
|
[Ankyrin repeat family protein] |
3.877 |
0.363 |
| 165 |
AT2G40095 |
AT2G40095
|
[Alpha/beta hydrolase related protein] |
3.877 |
no data |
| 166 |
AT5G22630 |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
3.876 |
-0.045 |
| 167 |
AT1G17750 |
AT1G17750
|
AtPEPR2, PEP1 RECEPTOR 2, PEPR2, PEP1 receptor 2 |
3.875 |
-0.080 |
| 168 |
AT5G54940 |
AT5G54940
|
[Translation initiation factor SUI1 family protein] |
3.874 |
0.004 |
| 169 |
AT5G47850 |
AT5G47850
|
CCR4, CRINKLY4 related 4 |
3.868 |
0.311 |
| 170 |
AT5G07990 |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
3.866 |
-0.010 |
| 171 |
AT2G32150 |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
3.865 |
-0.233 |
| 172 |
AT4G34150 |
AT4G34150
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
3.864 |
0.146 |
| 173 |
AT1G24140 |
AT1G24140
|
[Matrixin family protein] |
3.862 |
0.531 |
| 174 |
AT3G25795 |
AT3G25795
|
TAS4, trans acting siRNA 4 |
3.860 |
no data |
| 175 |
AT5G57220 |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
3.859 |
0.309 |
| 176 |
AT4G29290 |
AT4G29290
|
LCR26, low-molecular-weight cysteine-rich 26 |
3.846 |
no data |
| 177 |
AT3G30775 |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
3.841 |
-0.270 |
| 178 |
AT4G01250 |
AT4G01250
|
AtWRKY22, WRKY22 |
3.836 |
0.446 |
| 179 |
AT2G18560 |
AT2G18560
|
[UDP-Glycosyltransferase superfamily protein] |
3.836 |
no data |
| 180 |
AT1G66760 |
AT1G66760
|
[MATE efflux family protein] |
3.833 |
0.009 |
| 181 |
AT2G22500 |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
3.822 |
0.185 |
| 182 |
AT5G22380 |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
3.816 |
0.125 |
| 183 |
AT4G23220 |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
3.812 |
0.189 |
| 184 |
AT5G41740 |
AT5G41740
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
3.808 |
no data |
| 185 |
AT1G64065 |
AT1G64065
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
3.806 |
no data |
| 186 |
AT1G20310 |
AT1G20310
|
unknown |
3.801 |
0.492 |
| 187 |
AT5G13320 |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
3.797 |
0.289 |
| 188 |
AT3G07195 |
AT3G07195
|
[RPM1-interacting protein 4 (RIN4) family protein] |
3.790 |
no data |
| 189 |
AT4G29720 |
AT4G29720
|
ATPAO5, polyamine oxidase 5, PAO5, polyamine oxidase 5 |
3.789 |
-0.102 |
| 190 |
AT3G61190 |
AT3G61190
|
BAP1, BON association protein 1 |
3.787 |
0.298 |
| 191 |
AT1G21120 |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
3.787 |
0.413 |
| 192 |
AT2G26530 |
AT2G26530
|
AR781 |
3.785 |
0.132 |
| 193 |
AT3G16510 |
AT3G16510
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
3.783 |
no data |
| 194 |
AT1G30730 |
AT1G30730
|
[FAD-binding Berberine family protein] |
3.778 |
no data |
| 195 |
AT5G05300 |
AT5G05300
|
unknown |
3.778 |
0.268 |
| 196 |
AT5G05250 |
AT5G05250
|
unknown |
3.769 |
-0.201 |
| 197 |
AT5G58680 |
AT5G58680
|
[ARM repeat superfamily protein] |
3.763 |
0.124 |
| 198 |
AT3G04640 |
AT3G04640
|
[glycine-rich protein] |
3.763 |
0.087 |
| 199 |
AT3G44860 |
AT3G44860
|
FAMT, farnesoic acid carboxyl-O-methyltransferase |
3.747 |
no data |
| 200 |
AT5G16360 |
AT5G16360
|
[NC domain-containing protein-related] |
3.745 |
no data |
| 201 |
AT3G54150 |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
3.745 |
0.490 |
| 202 |
AT3G22910 |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
3.741 |
-0.047 |
| 203 |
AT3G44300 |
AT3G44300
|
AtNIT2, nitrilase 2, NIT2, nitrilase 2 |
3.732 |
no data |
| 204 |
AT5G57010 |
AT5G57010
|
[calmodulin-binding family protein] |
3.730 |
no data |
| 205 |
AT1G63860 |
AT1G63860
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
3.727 |
no data |
| 206 |
AT1G13609 |
AT1G13609
|
[Defensin-like (DEFL) family protein] |
3.724 |
no data |
| 207 |
AT1G18740 |
AT1G18740
|
unknown |
3.716 |
0.102 |
| 208 |
AT1G27730 |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
3.716 |
0.206 |
| 209 |
AT1G80120 |
AT1G80120
|
unknown |
3.707 |
no data |
| 210 |
AT3G25590 |
AT3G25590
|
unknown |
3.707 |
no data |
| 211 |
AT2G25625 |
AT2G25625
|
unknown |
3.697 |
-0.038 |
| 212 |
AT1G68690 |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
3.694 |
0.385 |
| 213 |
AT4G23810 |
AT4G23810
|
ATWRKY53, WRKY53 |
3.694 |
0.286 |
| 214 |
AT1G56060 |
AT1G56060
|
unknown |
3.690 |
0.285 |
| 215 |
AT3G17110 |
AT3G17110
|
[pseudogene] |
3.683 |
0.014 |
| 216 |
AT1G28190 |
AT1G28190
|
unknown |
3.666 |
0.299 |
| 217 |
AT4G21680 |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
3.660 |
0.055 |
| 218 |
AT5G17980 |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
3.658 |
-0.000 |
| 219 |
AT1G61820 |
AT1G61820
|
BGLU46, beta glucosidase 46 |
3.658 |
0.059 |
| 220 |
AT5G63130 |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
3.652 |
0.157 |
| 221 |
AT1G56540 |
AT1G56540
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
3.651 |
0.118 |
| 222 |
AT5G64260 |
AT5G64260
|
EXL2, EXORDIUM like 2 |
3.650 |
-0.056 |
| 223 |
AT1G20510 |
AT1G20510
|
OPCL1, OPC-8:0 CoA ligase1 |
3.646 |
0.150 |
| 224 |
AT4G30270 |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
3.646 |
-0.181 |
| 225 |
AT4G36010 |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
3.639 |
-0.149 |
| 226 |
AT4G24380 |
AT4G24380
|
[INVOLVED IN: 10-formyltetrahydrofolate biosynthetic process, folic acid and derivative biosynthetic process] |
3.636 |
0.084 |
| 227 |
AT5G63450 |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
3.635 |
-0.015 |
| 228 |
AT1G21240 |
AT1G21240
|
WAK3, wall associated kinase 3 |
3.632 |
0.032 |
| 229 |
AT2G36090 |
AT2G36090
|
[F-box family protein] |
3.632 |
0.252 |
| 230 |
AT1G61610 |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
3.631 |
-0.071 |
| 231 |
AT4G37370 |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
3.631 |
0.004 |
| 232 |
AT4G21390 |
AT4G21390
|
B120 |
3.629 |
0.421 |
| 233 |
AT2G01300 |
AT2G01300
|
unknown |
3.627 |
0.108 |
| 234 |
AT1G16130 |
AT1G16130
|
WAKL2, wall associated kinase-like 2 |
3.621 |
0.520 |
| 235 |
AT4G11000 |
AT4G11000
|
[Ankyrin repeat family protein] |
3.607 |
-0.041 |
| 236 |
AT5G22250 |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
3.603 |
0.293 |
| 237 |
AT5G25930 |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
3.603 |
0.359 |
| 238 |
AT5G59820 |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
3.600 |
0.267 |
| 239 |
AT2G32030 |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
3.600 |
0.357 |
| 240 |
AT4G23200 |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
3.600 |
0.120 |
| 241 |
AT4G36900 |
AT4G36900
|
DEAR4, DREB AND EAR MOTIF PROTEIN 4, RAP2.10, related to AP2 10 |
3.598 |
0.010 |
| 242 |
AT4G20830 |
AT4G20830
|
[FAD-binding Berberine family protein] |
3.596 |
0.191 |
| 243 |
AT3G49580 |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
3.595 |
0.038 |
| 244 |
AT1G18710 |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
3.583 |
-0.222 |
| 245 |
AT2G41100 |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
3.581 |
0.119 |
| 246 |
AT1G16030 |
AT1G16030
|
Hsp70b, heat shock protein 70B |
3.580 |
0.020 |
| 247 |
AT4G08040 |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
3.580 |
0.034 |
| 248 |
AT4G22780 |
AT4G22780
|
ACR7, ACT domain repeat 7 |
3.579 |
0.345 |
| 249 |
AT4G30430 |
AT4G30430
|
TET9, tetraspanin9 |
3.577 |
0.064 |
| 250 |
AT1G20823 |
AT1G20823
|
[RING/U-box superfamily protein] |
3.570 |
-0.028 |
| 251 |
AT4G27765 |
AT4G27765
|
miR828, MIR828A, microRNA828A |
3.567 |
no data |
| 252 |
AT5G40020 |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
3.567 |
-0.002 |
| 253 |
AT3G29670 |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
3.551 |
-0.015 |
| 254 |
AT3G57450 |
AT3G57450
|
unknown |
3.551 |
0.254 |
| 255 |
AT5G67340 |
AT5G67340
|
[ARM repeat superfamily protein] |
3.549 |
0.503 |
| 256 |
AT3G60220 |
AT3G60220
|
ATL4, TOXICOS EN LEVADURA 4, TL4, TOXICOS EN LEVADURA 4 |
3.538 |
-0.003 |
| 257 |
AT1G15010 |
AT1G15010
|
unknown |
3.537 |
-0.161 |
| 258 |
AT1G22810 |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
3.527 |
0.342 |
| 259 |
AT1G67481 |
AT1G67481
|
MIR839A, microRNA839A |
3.527 |
no data |
| 260 |
AT2G17660 |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
3.526 |
-0.088 |
| 261 |
AT3G22160 |
AT3G22160
|
JAV1, jasmonate-associated VQ motif gene 1 |
3.523 |
0.249 |
| 262 |
AT5G64310 |
AT5G64310
|
AGP1, arabinogalactan protein 1, ATAGP1 |
3.518 |
0.054 |
| 263 |
AT5G13190 |
AT5G13190
|
AtGILP, GILP, GSH-induced LITAF domain protein |
3.518 |
0.094 |
| 264 |
AT4G24570 |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
3.516 |
0.288 |
| 265 |
AT2G15390 |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
3.511 |
0.528 |
| 266 |
AT4G39190 |
AT4G39190
|
unknown |
3.511 |
-0.026 |
| 267 |
AT4G00700 |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
3.509 |
-0.047 |
| 268 |
AT4G23215 |
AT4G23215
|
|
3.507 |
no data |
| 269 |
AT1G12020 |
AT1G12020
|
unknown |
3.506 |
-0.161 |
| 270 |
AT4G21850 |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
3.504 |
0.169 |
| 271 |
AT1G07135 |
AT1G07135
|
[glycine-rich protein] |
3.503 |
no data |
| 272 |
AT5G50800 |
AT5G50800
|
AtSWEET13, SWEET13 |
3.502 |
-0.153 |
| 273 |
AT5G38700 |
AT5G38700
|
unknown |
3.498 |
-0.025 |
| 274 |
AT1G32170 |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
3.498 |
0.087 |
| 275 |
AT5G11650 |
AT5G11650
|
[alpha/beta-Hydrolases superfamily protein] |
3.495 |
0.080 |
| 276 |
AT3G24420 |
AT3G24420
|
[alpha/beta-Hydrolases superfamily protein] |
3.492 |
no data |
| 277 |
AT1G60190 |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
3.492 |
-0.062 |
| 278 |
AT1G24145 |
AT1G24145
|
unknown |
3.480 |
no data |
| 279 |
AT1G07000 |
AT1G07000
|
ATEXO70B2, exocyst subunit exo70 family protein B2, EXO70B2, exocyst subunit exo70 family protein B2 |
3.480 |
0.307 |
| 280 |
AT1G13670 |
AT1G13670
|
unknown |
3.469 |
-0.105 |
| 281 |
AT5G59550 |
AT5G59550
|
unknown |
3.464 |
0.141 |
| 282 |
AT2G27660 |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
3.463 |
0.481 |
| 283 |
AT5G19230 |
AT5G19230
|
[Glycoprotein membrane precursor GPI-anchored] |
3.463 |
no data |
| 284 |
AT1G23710 |
AT1G23710
|
unknown |
3.450 |
0.336 |
| 285 |
AT3G54020 |
AT3G54020
|
AtIPCS1, Arabidopsis Inositol phosphorylceramide synthase 1 |
3.448 |
no data |
| 286 |
AT2G03240 |
AT2G03240
|
[EXS (ERD1/XPR1/SYG1) family protein] |
3.443 |
-0.111 |
| 287 |
AT2G45680 |
AT2G45680
|
TCP9, TCP domain protein 9 |
3.441 |
0.000 |
| 288 |
AT5G46295 |
AT5G46295
|
unknown |
3.441 |
0.075 |
| 289 |
AT2G32140 |
AT2G32140
|
[transmembrane receptors] |
3.431 |
0.113 |
| 290 |
AT1G57990 |
AT1G57990
|
ATPUP18, purine permease 18, PUP18, purine permease 18 |
3.430 |
no data |
| 291 |
AT4G33050 |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
3.427 |
0.291 |
| 292 |
AT3G15210 |
AT3G15210
|
ATERF-4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ATERF4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ERF4, ethylene responsive element binding factor 4, RAP2.5, RELATED TO AP2 5 |
3.424 |
0.089 |
| 293 |
AT4G23190 |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
3.424 |
0.547 |
| 294 |
AT5G46080 |
AT5G46080
|
[Protein kinase superfamily protein] |
3.420 |
0.665 |
| 295 |
AT5G53730 |
AT5G53730
|
NHL26, NDR1/HIN1-like 26 |
3.419 |
no data |
| 296 |
AT2G28400 |
AT2G28400
|
unknown |
3.416 |
-0.012 |
| 297 |
AT5G39670 |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
3.413 |
0.383 |
| 298 |
AT5G46050 |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
3.412 |
0.237 |
| 299 |
AT5G45820 |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
3.412 |
-0.140 |
| 300 |
AT3G50260 |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
3.405 |
0.189 |
| 301 |
ATCG01160 |
ATCG01160
|
RRN5S, ribosomal RNA5S |
-6.597 |
no data |
| 302 |
AT5G25090 |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-3.937 |
-0.012 |
| 303 |
AT2G40610 |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-3.401 |
-0.304 |
| 304 |
ATCG00960 |
ATCG00960
|
RRN4.5S.1, ribosomal RNA4.5S |
-3.259 |
no data |
| 305 |
AT1G49870 |
AT1G49870
|
unknown |
-3.020 |
-0.033 |
| 306 |
AT2G40670 |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-2.993 |
-0.026 |
| 307 |
AT3G18773 |
AT3G18773
|
[RING/U-box superfamily protein] |
-2.859 |
no data |
| 308 |
AT5G24570 |
AT5G24570
|
unknown |
-2.622 |
-0.091 |
| 309 |
AT1G07050 |
AT1G07050
|
[CCT motif family protein] |
-2.540 |
-0.340 |
| 310 |
AT2G01020 |
AT2G01020
|
[5.8SrRNA] |
-2.462 |
no data |
| 311 |
AT1G73830 |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-2.437 |
0.044 |
| 312 |
AT1G60810 |
AT1G60810
|
ACLA-2, ATP-citrate lyase A-2 |
-2.385 |
0.001 |
| 313 |
AT1G65450 |
AT1G65450
|
GLC, GLAUCE |
-2.359 |
-0.072 |
| 314 |
AT3G51280 |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-2.314 |
-0.124 |
| 315 |
AT3G01680 |
AT3G01680
|
AtSEOR1, Arabidopsis thaliana sieve element occlusion-related 1, SEOb, sieve element occlusion b, SEOR1, Sieve-Element-Occlusion-Related 1 |
-2.310 |
-0.029 |
| 316 |
AT3G26960 |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-2.216 |
-0.210 |
| 317 |
AT1G53520 |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-2.177 |
-0.220 |
| 318 |
AT5G56120 |
AT5G56120
|
unknown |
-2.116 |
no data |
| 319 |
AT5G35260 |
AT5G35260
|
[similar to replication protein-related [Arabidopsis thaliana] (TAIR:AT4G07440.1)] |
-2.106 |
0.007 |
| 320 |
AT4G02850 |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-2.098 |
-0.219 |
| 321 |
AT1G80050 |
AT1G80050
|
APT2, adenine phosphoribosyl transferase 2, ATAPT2, PHT1.1 |
-2.042 |
-0.187 |
| 322 |
AT5G56550 |
AT5G56550
|
ATOXS3, OXIDATIVE STRESS 3, OXS3, OXIDATIVE STRESS 3 |
-2.042 |
no data |
| 323 |
AT2G33920 |
AT2G33920
|
[tRNA-Pro (anticodon: AGG)] |
-2.001 |
no data |
| 324 |
AT5G44430 |
AT5G44430
|
PDF1.2c, plant defensin 1.2C |
-1.985 |
no data |
| 325 |
AT2G26760 |
AT2G26760
|
CYCB1;4, Cyclin B1;4 |
-1.985 |
0.019 |
| 326 |
AT5G65890 |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-1.984 |
-0.115 |
| 327 |
AT2G39850 |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-1.977 |
-0.231 |
| 328 |
AT4G12320 |
AT4G12320
|
CYP706A6, cytochrome P450, family 706, subfamily A, polypeptide 6 |
-1.955 |
no data |
| 329 |
AT1G06080 |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-1.947 |
0.019 |
| 330 |
AT4G31840 |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-1.939 |
-0.180 |
| 331 |
AT1G52340 |
AT1G52340
|
ABA2, ABA DEFICIENT 2, ATABA2, ARABIDOPSIS THALIANA ABA DEFICIENT 2, ATSDR1, SHORT-CHAIN DEHYDROGENASE/REDUCTASE 1, GIN1, GLUCOSE INSENSITIVE 1, ISI4, IMPAIRED SUCROSE INDUCTION 4, SDR1, SHORT-CHAIN DEHYDROGENASE REDUCTASE 1, SIS4, SUGAR-INSENSITIVE 4, SRE1, SALT RESISTANT 1 |
-1.907 |
-0.093 |
| 332 |
AT1G03780 |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-1.907 |
-0.054 |
| 333 |
AT3G16670 |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-1.901 |
0.027 |
| 334 |
AT1G76135 |
AT1G76135
|
MIR394B, microRNA394B |
-1.860 |
no data |
| 335 |
AT2G32860 |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-1.851 |
-0.013 |
| 336 |
AT5G33370 |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.847 |
0.058 |
| 337 |
AT2G32160 |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.835 |
-0.058 |
| 338 |
AT1G01900 |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
-1.823 |
-0.209 |
| 339 |
AT1G26945 |
AT1G26945
|
KDR, KIDARI, PRE6, PACLOBUTRAZOL RESISTANCE 6 |
-1.823 |
no data |
| 340 |
AT3G10525 |
AT3G10525
|
LGO, LOSS OF GIANT CELLS FROM ORGANS, SMR1, SIAMESE RELATED 1 |
-1.822 |
-0.163 |
| 341 |
AT4G34630 |
AT4G34630
|
unknown |
-1.818 |
0.042 |
| 342 |
AT2G02850 |
AT2G02850
|
ARPN, plantacyanin |
-1.815 |
0.155 |
| 343 |
AT5G45950 |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.813 |
-0.101 |
| 344 |
AT5G48900 |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-1.802 |
-0.351 |
| 345 |
AT1G10640 |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-1.787 |
-0.119 |
| 346 |
AT1G02205 |
AT1G02205
|
CER1, ECERIFERUM 1, CER22, ECERIFERUM 22 |
-1.786 |
-0.130 |
| 347 |
AT1G18370 |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-1.770 |
-0.047 |
| 348 |
AT3G04140 |
AT3G04140
|
[Ankyrin repeat family protein] |
-1.765 |
no data |
| 349 |
AT1G60590 |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-1.759 |
-0.411 |
| 350 |
AT1G01600 |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-1.742 |
0.055 |
| 351 |
AT4G34730 |
AT4G34730
|
RBF1, RbfA domain-containing protein 1 |
-1.740 |
-0.217 |
| 352 |
AT1G74670 |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-1.740 |
-0.378 |
| 353 |
AT1G27120 |
AT1G27120
|
[Galactosyltransferase family protein] |
-1.722 |
-0.142 |
| 354 |
AT4G35350 |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-1.713 |
-0.036 |
| 355 |
AT5G44420 |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-1.704 |
0.040 |
| 356 |
AT1G67865 |
AT1G67865
|
unknown |
-1.694 |
0.033 |
| 357 |
AT3G62550 |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-1.681 |
-0.202 |
| 358 |
AT1G19371 |
AT1G19371
|
MIR169H, microRNA169H |
-1.678 |
no data |
| 359 |
AT4G15830 |
AT4G15830
|
[ARM repeat superfamily protein] |
-1.677 |
-0.223 |
| 360 |
AT5G58770 |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-1.666 |
-0.111 |
| 361 |
AT5G63180 |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-1.665 |
-0.117 |
| 362 |
AT2G30180 |
AT2G30180
|
[tRNA-Gly (anticodon: GCC)] |
-1.665 |
no data |
| 363 |
AT3G18110 |
AT3G18110
|
EMB1270, embryo defective 1270 |
-1.664 |
-0.254 |
| 364 |
AT4G28780 |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.658 |
-0.330 |
| 365 |
AT2G01010 |
AT2G01010
|
[18SrRNA] |
-1.644 |
no data |
| 366 |
AT4G12830 |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-1.634 |
-0.298 |
| 367 |
AT4G09520 |
AT4G09520
|
[Cofactor-independent phosphoglycerate mutase] |
-1.631 |
-0.015 |
| 368 |
AT4G26530 |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-1.625 |
-0.199 |
| 369 |
AT1G11850 |
AT1G11850
|
unknown |
-1.616 |
-0.150 |
| 370 |
AT4G27595 |
AT4G27595
|
unknown |
-1.597 |
no data |
| 371 |
AT5G46240 |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-1.591 |
0.100 |
| 372 |
AT4G18970 |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.583 |
-0.182 |
| 373 |
AT1G19950 |
AT1G19950
|
HVA22H, HVA22-like protein H (ATHVA22H) |
-1.575 |
-0.186 |
| 374 |
AT1G52770 |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-1.574 |
-0.212 |
| 375 |
AT5G25130 |
AT5G25130
|
CYP71B12, cytochrome P450, family 71, subfamily B, polypeptide 12 |
-1.574 |
-0.054 |
| 376 |
AT5G63780 |
AT5G63780
|
SHA1, shoot apical meristem arrest 1 |
-1.567 |
-0.162 |
| 377 |
AT2G40100 |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-1.562 |
-0.009 |
| 378 |
AT5G57170 |
AT5G57170
|
[Tautomerase/MIF superfamily protein] |
-1.560 |
-0.294 |
| 379 |
AT2G42610 |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-1.547 |
-0.073 |
| 380 |
AT1G14410 |
AT1G14410
|
ATWHY1, A. THALIANA WHIRLY 1, PTAC1, WHY1, WHIRLY 1 |
-1.541 |
-0.089 |
| 381 |
AT5G25460 |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-1.531 |
-0.148 |
| 382 |
AT1G67080 |
AT1G67080
|
ABA4, abscisic acid (ABA)-deficient 4 |
-1.527 |
-0.128 |
| 383 |
AT2G36885 |
AT2G36885
|
unknown |
-1.524 |
-0.076 |
| 384 |
AT1G48610 |
AT1G48610
|
[AT hook motif-containing protein] |
-1.518 |
-0.041 |
| 385 |
AT4G04840 |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-1.509 |
-0.171 |
| 386 |
AT1G72250 |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-1.479 |
-0.082 |
| 387 |
AT1G44110 |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-1.477 |
0.013 |
| 388 |
AT1G79460 |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-1.476 |
-0.262 |
| 389 |
AT2G05160 |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-1.476 |
-0.118 |
| 390 |
AT1G69530 |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-1.475 |
0.129 |
| 391 |
AT2G19170 |
AT2G19170
|
SLP3, subtilisin-like serine protease 3 |
-1.439 |
-0.196 |
| 392 |
AT2G03310 |
AT2G03310
|
unknown |
-1.439 |
-0.085 |
| 393 |
AT5G51720 |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-1.432 |
-0.133 |
| 394 |
AT1G71790 |
AT1G71790
|
AtCPB, CPB, capping protein B |
-1.431 |
-0.034 |
| 395 |
AT3G62820 |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.408 |
-0.028 |
| 396 |
AT5G41490 |
AT5G41490
|
[F-box associated ubiquitination effector family protein] |
-1.403 |
-0.008 |
| 397 |
AT1G18250 |
AT1G18250
|
ATLP-1 |
-1.395 |
-0.028 |
| 398 |
AT5G49740 |
AT5G49740
|
ATFRO7, FERRIC REDUCTION OXIDASE 7, FRO7, ferric reduction oxidase 7 |
-1.388 |
no data |
| 399 |
AT2G04063 |
AT2G04063
|
[glycine-rich protein] |
-1.383 |
0.063 |
| 400 |
AT4G32460 |
AT4G32460
|
unknown |
-1.383 |
-0.010 |
| 401 |
AT5G55620 |
AT5G55620
|
unknown |
-1.381 |
-0.098 |
| 402 |
AT1G61350 |
AT1G61350
|
[ARM repeat superfamily protein] |
-1.381 |
0.122 |
| 403 |
AT5G17670 |
AT5G17670
|
[alpha/beta-Hydrolases superfamily protein] |
-1.379 |
-0.113 |
| 404 |
AT1G66130 |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.343 |
-0.278 |
| 405 |
AT4G39330 |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-1.340 |
-0.009 |
| 406 |
AT2G28790 |
AT2G28790
|
[Pathogenesis-related thaumatin superfamily protein] |
-1.326 |
0.029 |
| 407 |
AT4G00780 |
AT4G00780
|
[TRAF-like family protein] |
-1.326 |
-0.083 |
| 408 |
AT2G25060 |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-1.325 |
-0.039 |
| 409 |
AT3G52380 |
AT3G52380
|
CP33, chloroplast RNA-binding protein 33, PDE322, PIGMENT DEFECTIVE 322 |
-1.319 |
-0.092 |
| 410 |
AT4G22200 |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-1.308 |
-0.285 |
| 411 |
AT4G38860 |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-1.295 |
-0.256 |
| 412 |
AT3G62060 |
AT3G62060
|
[Pectinacetylesterase family protein] |
-1.295 |
-0.119 |
| 413 |
AT3G56160 |
AT3G56160
|
[Sodium Bile acid symporter family] |
-1.295 |
-0.180 |
| 414 |
AT1G21460 |
AT1G21460
|
AtSWEET1, SWEET1 |
-1.293 |
-0.161 |
| 415 |
AT2G34900 |
AT2G34900
|
GTE01, GTE1, GLOBAL TRANSCRIPTION FACTOR GROUP E1, IMB1, IMBIBITION-INDUCIBLE 1 |
-1.291 |
-0.003 |
| 416 |
AT2G04032 |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-1.286 |
-0.074 |
| 417 |
AT3G49620 |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-1.275 |
0.213 |
| 418 |
AT2G45560 |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
-1.273 |
-0.181 |
| 419 |
AT1G02730 |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-1.252 |
-0.018 |
| 420 |
AT2G29290 |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.245 |
-0.254 |
| 421 |
AT2G04060 |
AT2G04060
|
[glycosyl hydrolase family 35 protein] |
-1.239 |
-0.003 |
| 422 |
AT5G65445 |
AT5G65445
|
[tRNA-Ser (anticodon: GCT)] |
-1.239 |
no data |
| 423 |
AT5G55740 |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-1.239 |
-0.390 |
| 424 |
AT1G31690 |
AT1G31690
|
[Copper amine oxidase family protein] |
-1.225 |
-0.295 |
| 425 |
AT1G51060 |
AT1G51060
|
HTA10, histone H2A 10 |
-1.218 |
-0.009 |
| 426 |
AT4G24350 |
AT4G24350
|
[Phosphorylase superfamily protein] |
-1.214 |
0.027 |
| 427 |
AT5G48910 |
AT5G48910
|
LPA66, LOW PSII ACCUMULATION 66 |
-1.210 |
-0.205 |
| 428 |
AT1G67870 |
AT1G67870
|
[glycine-rich protein] |
-1.208 |
-0.055 |
| 429 |
AT5G22580 |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-1.208 |
-0.131 |
| 430 |
AT5G53487 |
AT5G53487
|
[tRNA-Gly (anticodon: CCC)] |
-1.202 |
no data |
| 431 |
AT2G28150 |
AT2G28150
|
unknown |
-1.202 |
no data |
| 432 |
AT4G33660 |
AT4G33660
|
unknown |
-1.202 |
-0.164 |
| 433 |
AT3G62300 |
AT3G62300
|
unknown |
-1.201 |
-0.065 |
| 434 |
AT1G04800 |
AT1G04800
|
[glycine-rich protein] |
-1.200 |
-0.017 |
| 435 |
AT4G11175 |
AT4G11175
|
[Nucleic acid-binding, OB-fold-like protein] |
-1.193 |
-0.167 |
| 436 |
AT4G30620 |
AT4G30620
|
[Uncharacterised BCR, YbaB family COG0718] |
-1.192 |
-0.212 |
| 437 |
AT5G15230 |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
-1.189 |
-0.026 |
| 438 |
AT4G28660 |
AT4G28660
|
PSB28, photosystem II reaction center PSB28 protein |
-1.189 |
-0.147 |
| 439 |
AT3G09250 |
AT3G09250
|
[Nuclear transport factor 2 (NTF2) family protein] |
-1.182 |
-0.069 |
| 440 |
AT1G79050 |
AT1G79050
|
RECA1, homolog of bacterial RecA |
-1.181 |
-0.086 |
| 441 |
AT4G02290 |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-1.181 |
0.103 |
| 442 |
ATCG01130 |
ATCG01130
|
TIC214, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 214, YCF1.2 |
-1.178 |
no data |
| 443 |
AT2G28720 |
AT2G28720
|
[Histone superfamily protein] |
-1.166 |
-0.069 |
| 444 |
AT1G07790 |
AT1G07790
|
HTB1 |
-1.166 |
-0.086 |
| 445 |
AT1G01270 |
AT1G01270
|
[tRNA-Gln (anticodon: CTG)] |
-1.163 |
no data |
| 446 |
AT2G26010 |
AT2G26010
|
PDF1.3, plant defensin 1.3 |
-1.162 |
no data |
| 447 |
ATCG00780 |
ATCG00780
|
RPL14, ribosomal protein L14 |
-1.159 |
0.006 |
| 448 |
AT5G15780 |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
-1.158 |
-0.321 |
| 449 |
AT5G35220 |
AT5G35220
|
AMOS1, ammonium overly sensitive 1, EGY1, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN 1 |
-1.157 |
no data |
| 450 |
AT1G10960 |
AT1G10960
|
ATFD1, ferredoxin 1, FD1, ferredoxin 1 |
-1.157 |
-0.016 |
| 451 |
AT4G23800 |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-1.154 |
-0.070 |
| 452 |
AT3G14385 |
AT3G14385
|
MIR169F, microRNA169F |
-1.150 |
no data |
| 453 |
AT5G39210 |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-1.148 |
-0.333 |
| 454 |
AT3G12930 |
AT3G12930
|
[Lojap-related protein] |
-1.145 |
-0.241 |
| 455 |
AT3G27830 |
AT3G27830
|
RPL12, RIBOSOMAL PROTEIN L12, RPL12-A, ribosomal protein L12-A |
-1.135 |
no data |
| 456 |
AT3G54090 |
AT3G54090
|
FLN1, fructokinase-like 1 |
-1.135 |
-0.211 |
| 457 |
ATCG01120 |
ATCG01120
|
RPS15, chloroplast ribosomal protein S15 |
-1.134 |
0.044 |
| 458 |
AT3G01500 |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
-1.132 |
-0.016 |
| 459 |
AT1G75460 |
AT1G75460
|
[ATP-dependent protease La (LON) domain protein] |
-1.131 |
-0.128 |
| 460 |
AT3G26060 |
AT3G26060
|
ATPRX Q, PRXQ, peroxiredoxin Q |
-1.127 |
-0.081 |
| 461 |
AT3G42240 |
AT3G42240
|
unknown |
-1.126 |
-0.021 |
| 462 |
AT5G23940 |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-1.125 |
-0.095 |
| 463 |
AT5G44650 |
AT5G44650
|
AtCEST, Arabidopsis thaliana chloroplast protein-enhancing stress tolerance, CEST, chloroplast protein-enhancing stress tolerance, Y3IP1, Ycf3-interacting protein 1 |
-1.121 |
-0.094 |
| 464 |
AT5G64580 |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
-1.121 |
-0.192 |
| 465 |
AT3G17170 |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-1.115 |
-0.333 |
| 466 |
AT4G14230 |
AT4G14230
|
unknown |
-1.102 |
-0.003 |
| 467 |
AT1G19920 |
AT1G19920
|
APS2, ASA1, ATP SULFURYLASE ARABIDOPSIS 1 |
-1.100 |
-0.128 |
| 468 |
AT3G12610 |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-1.099 |
-0.205 |
| 469 |
AT2G21970 |
AT2G21970
|
SEP2, stress enhanced protein 2 |
-1.098 |
-0.076 |
| 470 |
AT4G13720 |
AT4G13720
|
[Inosine triphosphate pyrophosphatase family protein] |
-1.093 |
-0.043 |
| 471 |
AT5G20630 |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-1.092 |
-0.359 |
| 472 |
AT1G29530 |
AT1G29530
|
unknown |
-1.090 |
-0.201 |
| 473 |
AT5G08280 |
AT5G08280
|
HEMC, hydroxymethylbilane synthase, RUG1, RUGOSA 1 |
-1.088 |
-0.064 |
| 474 |
AT3G26200 |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-1.085 |
0.186 |
| 475 |
AT5G64770 |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-1.084 |
-0.157 |
| 476 |
AT5G51330 |
AT5G51330
|
DYAD, SWI1, SWITCH1 |
-1.082 |
no data |
| 477 |
AT3G16250 |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-1.077 |
-0.201 |
| 478 |
AT3G44990 |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-1.074 |
-0.317 |
| 479 |
AT1G12900 |
AT1G12900
|
GAPA-2, glyceraldehyde 3-phosphate dehydrogenase A subunit 2 |
-1.074 |
-0.074 |
| 480 |
AT3G52150 |
AT3G52150
|
PSRP2, plastid-specific ribosomal protein 2 |
-1.073 |
-0.155 |
| 481 |
AT4G09010 |
AT4G09010
|
APX4, ascorbate peroxidase 4, TL29, thylakoid lumen 29 |
-1.069 |
-0.093 |
| 482 |
AT4G16980 |
AT4G16980
|
[arabinogalactan-protein family] |
-1.066 |
-0.132 |
| 483 |
AT2G37640 |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-1.063 |
-0.032 |
| 484 |
AT2G40400 |
AT2G40400
|
unknown |
-1.059 |
-0.062 |
| 485 |
AT5G60963 |
AT5G60963
|
[tRNA-Ala (anticodon: CGC)] |
-1.058 |
no data |
| 486 |
ATCG00760 |
ATCG00760
|
RPL36, ribosomal protein L36 |
-1.057 |
-0.036 |
| 487 |
AT1G64670 |
AT1G64670
|
BDG1, BODYGUARD1, CED1, 9-CIS EPOXYCAROTENOID DIOXYGENASE DEFECTIVE 1 |
-1.057 |
-0.085 |
| 488 |
AT4G00895 |
AT4G00895
|
[ATPase, F1 complex, OSCP/delta subunit protein] |
-1.055 |
-0.136 |
| 489 |
AT4G34950 |
AT4G34950
|
[Major facilitator superfamily protein] |
-1.051 |
-0.065 |
| 490 |
AT1G14150 |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-1.051 |
-0.256 |
| 491 |
AT5G60210 |
AT5G60210
|
RIP5, ROP interactive partner 5 |
-1.047 |
-0.098 |
| 492 |
AT1G15980 |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
-1.047 |
-0.176 |
| 493 |
AT3G24590 |
AT3G24590
|
PLSP1, plastidic type i signal peptidase 1 |
-1.045 |
-0.101 |
| 494 |
AT5G10430 |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-1.045 |
-0.100 |
| 495 |
ATCG00160 |
ATCG00160
|
RPS2, ribosomal protein S2 |
-1.045 |
-0.002 |
| 496 |
AT5G38410 |
AT5G38410
|
RBCS3B, Rubisco small subunit 3B |
-1.041 |
no data |
| 497 |
AT5G38420 |
AT5G38420
|
RBCS2B, Rubisco small subunit 2B |
-1.040 |
no data |
| 498 |
AT5G57030 |
AT5G57030
|
LUT2, LUTEIN DEFICIENT 2 |
-1.040 |
-0.163 |
| 499 |
AT1G13930 |
AT1G13930
|
unknown |
-1.040 |
-0.040 |
| 500 |
AT2G40660 |
AT2G40660
|
[Nucleic acid-binding, OB-fold-like protein] |
-1.036 |
-0.039 |
| 501 |
AT5G40950 |
AT5G40950
|
RPL27, ribosomal protein large subunit 27 |
-1.035 |
-0.110 |
| 502 |
AT1G56050 |
AT1G56050
|
[GTP-binding protein-related] |
-1.035 |
-0.167 |
| 503 |
AT3G27060 |
AT3G27060
|
ATTSO2, TSO2, TSO MEANING 'UGLY' IN CHINESE 2 |
-1.029 |
-0.098 |
| 504 |
AT5G61160 |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-1.026 |
0.363 |
| 505 |
AT3G23890 |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-1.024 |
-0.030 |
| 506 |
AT3G09860 |
AT3G09860
|
unknown |
-1.024 |
no data |
| 507 |
AT5G51750 |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
-1.014 |
-0.139 |
| 508 |
AT5G67280 |
AT5G67280
|
RLK, receptor-like kinase |
-1.012 |
-0.159 |
| 509 |
ATCG00810 |
ATCG00810
|
RPL22, ribosomal protein L22 |
-1.012 |
-0.023 |
| 510 |
AT5G08000 |
AT5G08000
|
E13L3, glucan endo-1,3-beta-glucosidase-like protein 3, PDCB2, PLASMODESMATA CALLOSE-BINDING PROTEIN 2 |
-1.012 |
-0.322 |
| 511 |
AT3G22150 |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.010 |
-0.317 |
| 512 |
AT5G02120 |
AT5G02120
|
OHP, one helix protein, PDE335, PIGMENT DEFECTIVE 335 |
-1.010 |
-0.127 |
| 513 |
AT5G18570 |
AT5G18570
|
ATOBGC, ATOBGL, OBG-like protein, CPSAR1, chloroplastic SAR1, EMB269, EMBRYO DEFECTIVE 269, EMB3138, EMBRYO DEFECTIVE 3138 |
-1.008 |
-0.195 |
| 514 |
AT1G32080 |
AT1G32080
|
AtLrgB, LrgB |
-1.008 |
-0.086 |
| 515 |
ATCG00065 |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
-1.007 |
-0.220 |
| 516 |
AT5G63550 |
AT5G63550
|
[DEK domain-containing chromatin associated protein] |
-1.006 |
no data |
| 517 |
AT3G47650 |
AT3G47650
|
[DnaJ/Hsp40 cysteine-rich domain superfamily protein] |
-1.005 |
-0.039 |
| 518 |
AT3G26570 |
AT3G26570
|
ORF02, PHT2;1, phosphate transporter 2;1 |
-1.004 |
-0.119 |
| 519 |
AT4G38160 |
AT4G38160
|
pde191, pigment defective 191 |
-0.992 |
-0.273 |
| 520 |
AT3G55630 |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
-0.992 |
-0.230 |
| 521 |
AT4G31850 |
AT4G31850
|
PGR3, proton gradient regulation 3 |
-0.991 |
-0.239 |
| 522 |
AT5G38430 |
AT5G38430
|
RBCS1B, Rubisco small subunit 1B |
-0.990 |
no data |
| 523 |
AT5G48460 |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.990 |
-0.216 |
| 524 |
AT3G47070 |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
-0.988 |
-0.084 |
| 525 |
AT3G13470 |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
-0.987 |
-0.061 |
| 526 |
AT4G18240 |
AT4G18240
|
ATSS4, ARABIDOPSIS THALIANA STARCH SYNTHASE 4, SS4, starch synthase 4, SSIV, STARCH SYNTHASE 4 |
-0.986 |
-0.113 |
| 527 |
AT1G08540 |
AT1G08540
|
ABC1, ATSIG1, SIGMA FACTOR 1, ATSIG2, SIGMA FACTOR 2, SIG1, RNA POLYMERASE SIGMA SUBUNIT 1, SIG2, RNApolymerase sigma subunit 2, SIGA, SIGB, SIGMA FACTOR B |
-0.986 |
-0.193 |
| 528 |
AT4G34035 |
AT4G34035
|
[tRNA-Arg (anticodon: TCT)] |
-0.984 |
no data |
| 529 |
AT5G61590 |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.982 |
-0.038 |
| 530 |
AT1G12520 |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
-0.980 |
-0.056 |
| 531 |
AT4G34830 |
AT4G34830
|
MRL1, MATURATION OF RBCL 1, PDE346, PIGMENT DEFECTIVE 346 |
-0.978 |
-0.196 |
| 532 |
AT1G67750 |
AT1G67750
|
[Pectate lyase family protein] |
-0.978 |
-0.255 |
| 533 |
AT1G75690 |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
-0.977 |
-0.118 |
| 534 |
AT5G52970 |
AT5G52970
|
[thylakoid lumen 15.0 kDa protein] |
-0.975 |
-0.146 |
| 535 |
AT2G18940 |
AT2G18940
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.974 |
-0.192 |
| 536 |
ATCG00770 |
ATCG00770
|
RPS8, ribosomal protein S8 |
-0.971 |
-0.011 |
| 537 |
AT2G45770 |
AT2G45770
|
CPFTSY, FRD4, FERRIC CHELATE REDUCTASE DEFECTIVE 4 |
-0.968 |
-0.134 |
| 538 |
AT3G16910 |
AT3G16910
|
AAE7, acyl-activating enzyme 7, ACN1, ACETATE NON-UTILIZING 1 |
-0.966 |
-0.093 |
| 539 |
AT3G48500 |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
-0.966 |
-0.341 |
| 540 |
AT3G10690 |
AT3G10690
|
GYRA, DNA GYRASE A |
-0.965 |
-0.027 |
| 541 |
AT3G54600 |
AT3G54600
|
DJ-1f, DJ1F |
-0.964 |
no data |
| 542 |
AT1G48350 |
AT1G48350
|
EMB3105, EMBRYO DEFECTIVE 3105 |
-0.961 |
-0.132 |
| 543 |
AT5G19540 |
AT5G19540
|
unknown |
-0.961 |
-0.071 |
| 544 |
AT1G74070 |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.958 |
-0.228 |
| 545 |
AT4G25050 |
AT4G25050
|
ACP4, acyl carrier protein 4 |
-0.957 |
-0.067 |
| 546 |
ATCG00170 |
ATCG00170
|
RPOC2 |
-0.946 |
-0.069 |
| 547 |
AT1G14840 |
AT1G14840
|
ATMAP70-4, microtubule-associated proteins 70-4, MAP70-4, microtubule-associated proteins 70-4 |
-0.946 |
-0.156 |
| 548 |
AT4G17560 |
AT4G17560
|
[Ribosomal protein L19 family protein] |
-0.945 |
-0.148 |
| 549 |
AT1G24020 |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.945 |
-0.097 |
| 550 |
AT1G32550 |
AT1G32550
|
FdC2, ferredoxin C 2 |
-0.944 |
-0.098 |
| 551 |
AT5G23920 |
AT5G23920
|
unknown |
-0.943 |
-0.148 |
| 552 |
AT1G09750 |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.943 |
-0.397 |
| 553 |
AT1G62380 |
AT1G62380
|
ACO2, ACC oxidase 2, ATACO2 |
-0.941 |
-0.020 |
| 554 |
AT1G76080 |
AT1G76080
|
ATCDSP32, ARABIDOPSIS THALIANA CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD, CDSP32, chloroplastic drought-induced stress protein of 32 kD |
-0.939 |
-0.011 |
| 555 |
AT4G00370 |
AT4G00370
|
ANTR2, PHT4;4, anion transporter 2 |
-0.938 |
-0.202 |
| 556 |
AT1G29070 |
AT1G29070
|
[Ribosomal protein L34] |
-0.938 |
-0.078 |
| 557 |
AT4G24750 |
AT4G24750
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.938 |
-0.157 |
| 558 |
AT2G33850 |
AT2G33850
|
unknown |
-0.936 |
-0.069 |
| 559 |
AT5G06290 |
AT5G06290
|
2-Cys Prx B, 2-cysteine peroxiredoxin B, 2CPB, 2-CYS PEROXIREDOXIN B |
-0.934 |
-0.065 |
| 560 |
AT5G59870 |
AT5G59870
|
HTA6, histone H2A 6 |
-0.933 |
-0.031 |
| 561 |
AT1G03130 |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
-0.932 |
-0.023 |
| 562 |
AT2G35780 |
AT2G35780
|
scpl26, serine carboxypeptidase-like 26 |
-0.931 |
-0.218 |
| 563 |
ATCG00370 |
ATCG00370
|
TRNS.3 |
-0.929 |
no data |
| 564 |
AT1G12800 |
AT1G12800
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.927 |
-0.139 |
| 565 |
AT2G40080 |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.926 |
0.048 |
| 566 |
AT5G64040 |
AT5G64040
|
PSAN |
-0.925 |
-0.048 |
| 567 |
AT3G48360 |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.925 |
0.053 |
| 568 |
AT2G33620 |
AT2G33620
|
AHL10, AT-hook motif nuclear localized protein 10 |
-0.924 |
0.031 |
| 569 |
AT1G35680 |
AT1G35680
|
RPL21C, chloroplast ribosomal protein L21 |
-0.922 |
-0.108 |
| 570 |
AT4G33580 |
AT4G33580
|
ATBCA5, A. THALIANA BETA CARBONIC ANHYDRASE 5, BCA5, beta carbonic anhydrase 5 |
-0.921 |
-0.091 |
| 571 |
AT1G75780 |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.921 |
-0.059 |
| 572 |
AT3G49500 |
AT3G49500
|
RDR6, RNA-dependent RNA polymerase 6, SDE1, SILENCING DEFECTIVE 1, SGS2, SUPPRESSOR OF GENE SILENCING 2 |
-0.918 |
-0.073 |
| 573 |
AT5G26650 |
AT5G26650
|
AGL36, AGAMOUS-like 36 |
-0.917 |
no data |
| 574 |
AT2G36830 |
AT2G36830
|
GAMMA-TIP, gamma tonoplast intrinsic protein, GAMMA-TIP1, GAMMA TONOPLAST INTRINSIC PROTEIN 1, TIP1;1, TONOPLAST INTRINSIC PROTEIN 1;1 |
-0.914 |
0.005 |
| 575 |
AT3G10020 |
AT3G10020
|
unknown |
-0.914 |
-0.036 |
| 576 |
AT5G07030 |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.913 |
-0.233 |
| 577 |
ATCG00040 |
ATCG00040
|
MATK, maturase K |
-0.912 |
-0.022 |
| 578 |
AT3G04790 |
AT3G04790
|
EMB3119, EMBRYO DEFECTIVE 3119 |
-0.912 |
-0.044 |
| 579 |
AT3G46740 |
AT3G46740
|
MAR1, MODIFIER OF ARG1 1, TOC75-III, translocon at the outer envelope membrane of chloroplasts 75-III |
-0.911 |
-0.043 |
| 580 |
AT1G60950 |
AT1G60950
|
ATFD2, FERREDOXIN 2, FD2, FED A |
-0.910 |
-0.031 |
| 581 |
AT3G49110 |
AT3G49110
|
ATPCA, PEROXIDASE CA, ATPRX33, PRX33, PEROXIDASE 33, PRXCA, peroxidase CA |
-0.909 |
no data |
| 582 |
AT4G21280 |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
-0.909 |
-0.058 |
| 583 |
ATCG00490 |
ATCG00490
|
RBCL |
-0.908 |
-0.091 |
| 584 |
AT2G18300 |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.908 |
-0.064 |
| 585 |
AT4G13170 |
AT4G13170
|
[Ribosomal protein L13 family protein] |
-0.907 |
-0.075 |
| 586 |
AT3G01440 |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.906 |
-0.377 |
| 587 |
AT1G33811 |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.905 |
-0.127 |
| 588 |
AT5G48360 |
AT5G48360
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.905 |
-0.157 |
| 589 |
ATCG01020 |
ATCG01020
|
RPL32, ribosomal protein L32 |
-0.903 |
0.014 |
| 590 |
ATCG01070 |
ATCG01070
|
NDHE |
-0.902 |
-0.073 |
| 591 |
AT1G68590 |
AT1G68590
|
PSRP3/1, plastid-specific ribosomal protein 3/1 |
-0.902 |
-0.183 |
| 592 |
AT3G15190 |
AT3G15190
|
PRPS20, plastid ribosomal protein S20 |
-0.900 |
-0.097 |
| 593 |
AT3G53190 |
AT3G53190
|
[Pectin lyase-like superfamily protein] |
-0.899 |
-0.165 |
| 594 |
ATCG00130 |
ATCG00130
|
ATPF |
-0.899 |
-0.021 |
| 595 |
AT5G66470 |
AT5G66470
|
[RNA binding] |
-0.897 |
-0.159 |
| 596 |
ATCG00790 |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.896 |
-0.037 |
| 597 |
AT5G17170 |
AT5G17170
|
ENH1, enhancer of sos3-1 |
-0.896 |
-0.110 |
| 598 |
AT5G55730 |
AT5G55730
|
FLA1, FASCICLIN-like arabinogalactan 1 |
-0.895 |
-0.231 |
| 599 |
AT5G13280 |
AT5G13280
|
AK, ASPARTATE KINASE, AK-LYS1, aspartate kinase 1, AK1, ASPARTATE KINASE 1 |
-0.895 |
0.046 |
| 600 |
AT1G17570 |
AT1G17570
|
[tRNA-Ser (anticodon: AGA)] |
-0.895 |
no data |