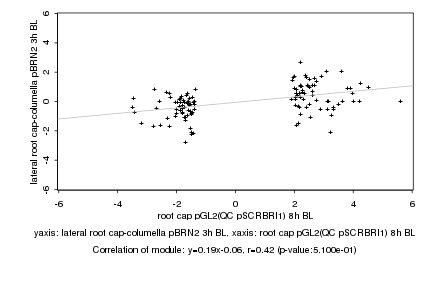
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root cap pGL2(QC pSCRBRI1) 8h BL |
Log2 signal ratio
lateral root cap-columella pBRN2 3h BL |
| 1 |
AT4G27160 |
AT4G27160
|
AT2S3, SESA3, seed storage albumin 3 |
5.592 |
0.035 |
| 2 |
AT1G44010 |
AT1G44010
|
unknown |
4.487 |
0.979 |
| 3 |
AT4G37780 |
AT4G37780
|
ATMYB87, MYB87, myb domain protein 87 |
4.238 |
1.258 |
| 4 |
AT3G45060 |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
4.202 |
0.035 |
| 5 |
AT2G04070 |
AT2G04070
|
[MATE efflux family protein] |
4.006 |
0.035 |
| 6 |
AT2G37810 |
AT2G37810
|
[Cysteine/Histidine-rich C1 domain family protein] |
3.962 |
0.566 |
| 7 |
AT4G03480 |
AT4G03480
|
[Ankyrin repeat family protein] |
3.891 |
0.935 |
| 8 |
AT2G37800 |
AT2G37800
|
[Cysteine/Histidine-rich C1 domain family protein] |
3.778 |
0.953 |
| 9 |
AT4G18195 |
AT4G18195
|
ATPUP8, ARABIDOPSIS THALIANA PURINE PERMEASE 8, PUP8, purine permease 8 |
3.610 |
0.035 |
| 10 |
AT5G39100 |
AT5G39100
|
GLP6, germin-like protein 6 |
3.583 |
2.077 |
| 11 |
AT4G14365 |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
3.496 |
-0.192 |
| 12 |
AT3G22231 |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
3.321 |
-0.353 |
| 13 |
AT1G48990 |
AT1G48990
|
[Oleosin family protein] |
3.311 |
-0.495 |
| 14 |
AT5G05390 |
AT5G05390
|
LAC12, laccase 12 |
3.252 |
-0.936 |
| 15 |
AT1G70260 |
AT1G70260
|
UMAMIT36, Usually multiple acids move in and out Transporters 36 |
3.204 |
-2.105 |
| 16 |
AT3G51200 |
AT3G51200
|
SAUR18, SMALL AUXIN UPREGULATED RNA 18 |
3.145 |
0.035 |
| 17 |
AT5G36920 |
AT5G36920
|
unknown |
3.126 |
-0.496 |
| 18 |
AT5G43290 |
AT5G43290
|
ATWRKY49, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 49, WRKY49, WRKY DNA-binding protein 49 |
3.126 |
0.035 |
| 19 |
AT1G33840 |
AT1G33840
|
unknown |
3.062 |
2.067 |
| 20 |
AT1G19250 |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
2.889 |
1.753 |
| 21 |
AT5G36925 |
AT5G36925
|
unknown |
2.859 |
-0.520 |
| 22 |
AT5G48430 |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
2.726 |
1.430 |
| 23 |
AT1G49860 |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
2.720 |
0.079 |
| 24 |
AT1G51913 |
AT1G51913
|
[LOCATED IN: endomembrane system] |
2.658 |
1.125 |
| 25 |
AT1G26420 |
AT1G26420
|
[FAD-binding Berberine family protein] |
2.655 |
1.620 |
| 26 |
AT3G55700 |
AT3G55700
|
[UDP-Glycosyltransferase superfamily protein] |
2.590 |
0.686 |
| 27 |
AT1G21320 |
AT1G21320
|
[nucleotide binding] |
2.585 |
0.442 |
| 28 |
AT3G20380 |
AT3G20380
|
[TRAF-like family protein] |
2.584 |
1.130 |
| 29 |
AT5G44568 |
AT5G44568
|
unknown |
2.537 |
-1.053 |
| 30 |
AT2G41100 |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
2.513 |
-0.141 |
| 31 |
AT4G18940 |
AT4G18940
|
[RNA ligase/cyclic nucleotide phosphodiesterase family protein] |
2.502 |
1.523 |
| 32 |
AT4G01970 |
AT4G01970
|
AtSTS, stachyose synthase, RS4, raffinose synthase 4, STS, stachyose synthase |
2.498 |
0.993 |
| 33 |
AT5G37490 |
AT5G37490
|
[ARM repeat superfamily protein] |
2.476 |
1.030 |
| 34 |
AT2G45360 |
AT2G45360
|
unknown |
2.446 |
1.138 |
| 35 |
AT4G12490 |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
2.412 |
-0.360 |
| 36 |
AT3G01175 |
AT3G01175
|
unknown |
2.400 |
1.674 |
| 37 |
AT1G26210 |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
2.367 |
1.776 |
| 38 |
AT4G14630 |
AT4G14630
|
GLP9, germin-like protein 9 |
2.332 |
0.592 |
| 39 |
AT1G49000 |
AT1G49000
|
unknown |
2.296 |
0.189 |
| 40 |
AT1G36640 |
AT1G36640
|
unknown |
2.276 |
0.620 |
| 41 |
AT3G50470 |
AT3G50470
|
HR3, homolog of RPW8 3, MLA10, INTRACELLULAR MILDEW A 10 |
2.267 |
0.796 |
| 42 |
AT1G65484 |
AT1G65484
|
unknown |
2.228 |
1.079 |
| 43 |
AT5G52710 |
AT5G52710
|
[Copper transport protein family] |
2.206 |
-0.831 |
| 44 |
AT3G51220 |
AT3G51220
|
unknown |
2.196 |
0.300 |
| 45 |
AT3G27095 |
AT3G27095
|
[pseudogene] |
2.194 |
1.047 |
| 46 |
AT1G22890 |
AT1G22890
|
unknown |
2.188 |
2.696 |
| 47 |
AT2G31540 |
AT2G31540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
2.179 |
1.102 |
| 48 |
AT1G09932 |
AT1G09932
|
[Phosphoglycerate mutase family protein] |
2.174 |
-0.373 |
| 49 |
AT3G62690 |
AT3G62690
|
ATL5, AtL5 |
2.149 |
0.561 |
| 50 |
AT1G29860 |
AT1G29860
|
ATWRKY71, WRKY DNA-BINDING PROTEIN 71, WRKY71, WRKY DNA-binding protein 71 |
2.148 |
0.566 |
| 51 |
AT5G42325 |
AT5G42325
|
[Transcription factor IIS protein] |
2.123 |
-1.492 |
| 52 |
AT2G18650 |
AT2G18650
|
MEE16, maternal effect embryo arrest 16 |
2.111 |
-0.332 |
| 53 |
AT5G19890 |
AT5G19890
|
[Peroxidase superfamily protein] |
2.082 |
0.431 |
| 54 |
AT4G04700 |
AT4G04700
|
CPK27, calcium-dependent protein kinase 27 |
2.065 |
0.549 |
| 55 |
AT4G14450 |
AT4G14450
|
unknown |
2.048 |
-1.625 |
| 56 |
AT5G41300 |
AT5G41300
|
[Receptor-like protein kinase-related family protein] |
2.045 |
0.584 |
| 57 |
AT1G21130 |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
2.044 |
0.883 |
| 58 |
AT3G50770 |
AT3G50770
|
CML41, calmodulin-like 41 |
2.020 |
0.145 |
| 59 |
AT2G39200 |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
2.018 |
-0.243 |
| 60 |
AT3G13610 |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
2.009 |
0.298 |
| 61 |
AT4G26260 |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
1.988 |
0.951 |
| 62 |
AT1G30040 |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
1.974 |
1.749 |
| 63 |
AT1G65486 |
AT1G65486
|
unknown |
1.963 |
1.653 |
| 64 |
AT3G50460 |
AT3G50460
|
HR2, homolog of RPW8 2 |
1.921 |
1.463 |
| 65 |
AT5G59930 |
AT5G59930
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.894 |
0.189 |
| 66 |
AT1G52342 |
AT1G52342
|
unknown |
-3.529 |
-0.399 |
| 67 |
ATCG00130 |
ATCG00130
|
ATPF |
-3.482 |
0.246 |
| 68 |
AT4G22217 |
AT4G22217
|
[Arabidopsis defensin-like protein] |
-3.429 |
-0.714 |
| 69 |
AT1G33100 |
AT1G33100
|
[MATE efflux family protein] |
-3.193 |
-1.483 |
| 70 |
AT3G25655 |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
-2.799 |
-1.640 |
| 71 |
AT5G59080 |
AT5G59080
|
unknown |
-2.754 |
0.850 |
| 72 |
AT4G15233 |
AT4G15233
|
ABCG42, ATP-binding cassette G42 |
-2.695 |
-0.466 |
| 73 |
AT1G26762 |
AT1G26762
|
unknown |
-2.593 |
0.035 |
| 74 |
AT1G33090 |
AT1G33090
|
[MATE efflux family protein] |
-2.578 |
-1.604 |
| 75 |
ATMG00690 |
ATMG00690
|
ORF240A |
-2.343 |
0.650 |
| 76 |
AT5G62210 |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-2.337 |
-1.121 |
| 77 |
AT2G25150 |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
-2.267 |
0.593 |
| 78 |
AT4G01680 |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-2.259 |
-1.660 |
| 79 |
ATCG00510 |
ATCG00510
|
PSAI, photsystem I subunit I |
-2.214 |
0.310 |
| 80 |
AT2G07708 |
AT2G07708
|
unknown |
-2.053 |
-0.048 |
| 81 |
AT5G48010 |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-2.042 |
-0.984 |
| 82 |
AT2G01021 |
AT2G01021
|
unknown |
-2.031 |
-0.522 |
| 83 |
AT3G49580 |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-2.014 |
-0.769 |
| 84 |
AT5G59760 |
AT5G59760
|
unknown |
-1.993 |
-0.016 |
| 85 |
AT2G34925 |
AT2G34925
|
CLE42, CLAVATA3/ESR-RELATED 42 |
-1.928 |
0.178 |
| 86 |
AT5G13080 |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-1.900 |
-0.326 |
| 87 |
AT2G07719 |
AT2G07719
|
[Putative membrane lipoprotein] |
-1.894 |
-0.039 |
| 88 |
AT5G12270 |
AT5G12270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.891 |
-0.550 |
| 89 |
AT5G66170 |
AT5G66170
|
STR18, sulfurtransferase 18 |
-1.889 |
0.249 |
| 90 |
AT1G70880 |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.867 |
-0.562 |
| 91 |
AT1G58170 |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-1.832 |
0.390 |
| 92 |
ATCG00040 |
ATCG00040
|
MATK, maturase K |
-1.818 |
-0.429 |
| 93 |
AT4G11190 |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-1.809 |
-0.768 |
| 94 |
AT5G65300 |
AT5G65300
|
unknown |
-1.808 |
-0.700 |
| 95 |
AT2G22770 |
AT2G22770
|
NAI1 |
-1.802 |
-0.253 |
| 96 |
AT4G37620 |
AT4G37620
|
[similar to RNase H domain-containing protein [Arabidopsis thaliana] (TAIR:AT4G09490.1)] |
-1.799 |
-0.428 |
| 97 |
AT1G19050 |
AT1G19050
|
ARR7, response regulator 7 |
-1.791 |
0.111 |
| 98 |
ATCG00360 |
ATCG00360
|
YCF3 |
-1.762 |
-0.020 |
| 99 |
AT5G48000 |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-1.753 |
-0.341 |
| 100 |
AT2G17070 |
AT2G17070
|
unknown |
-1.749 |
-1.045 |
| 101 |
AT1G21520 |
AT1G21520
|
unknown |
-1.734 |
-0.008 |
| 102 |
AT3G62760 |
AT3G62760
|
ATGSTF13 |
-1.727 |
-2.761 |
| 103 |
AT1G64950 |
AT1G64950
|
CYP89A5, cytochrome P450, family 89, subfamily A, polypeptide 5 |
-1.711 |
-1.261 |
| 104 |
AT3G49570 |
AT3G49570
|
LSU3, RESPONSE TO LOW SULFUR 3 |
-1.701 |
-1.072 |
| 105 |
AT1G26770 |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
-1.675 |
0.429 |
| 106 |
AT1G70885 |
AT1G70885
|
[pseudogene] |
-1.653 |
-0.075 |
| 107 |
AT1G68620 |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-1.643 |
0.563 |
| 108 |
AT4G01670 |
AT4G01670
|
unknown |
-1.641 |
-0.928 |
| 109 |
AT5G47990 |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-1.638 |
-0.066 |
| 110 |
AT4G30270 |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
-1.622 |
-0.563 |
| 111 |
AT2G31980 |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-1.612 |
0.056 |
| 112 |
AT3G15030 |
AT3G15030
|
MEE35, maternal effect embryo arrest 35, TCP4, TCP family transcription factor 4 |
-1.610 |
-0.108 |
| 113 |
AT1G72490 |
AT1G72490
|
unknown |
-1.590 |
0.217 |
| 114 |
AT5G23820 |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-1.561 |
-0.222 |
| 115 |
AT5G57010 |
AT5G57010
|
[calmodulin-binding family protein] |
-1.555 |
-1.792 |
| 116 |
AT2G25450 |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.527 |
-0.664 |
| 117 |
AT1G75450 |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
-1.527 |
-0.162 |
| 118 |
AT5G38940 |
AT5G38940
|
[RmlC-like cupins superfamily protein] |
-1.523 |
-2.202 |
| 119 |
AT2G41312 |
AT2G41312
|
other RNA |
-1.517 |
-0.751 |
| 120 |
AT5G38020 |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.510 |
-2.084 |
| 121 |
AT5G26290 |
AT5G26290
|
[TRAF-like family protein] |
-1.507 |
-0.875 |
| 122 |
AT1G14960 |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.471 |
-0.685 |
| 123 |
ATMG00090 |
ATMG00090
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
-1.461 |
0.328 |
| 124 |
ATCG00790 |
ATCG00790
|
RPL16, ribosomal protein L16 |
-1.454 |
-0.020 |
| 125 |
AT3G30180 |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-1.436 |
-2.131 |
| 126 |
AT2G39310 |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
-1.417 |
-0.536 |
| 127 |
AT3G45680 |
AT3G45680
|
[Major facilitator superfamily protein] |
-1.412 |
0.067 |
| 128 |
AT1G10370 |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-1.400 |
-0.157 |
| 129 |
AT1G62975 |
AT1G62975
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-1.385 |
0.881 |