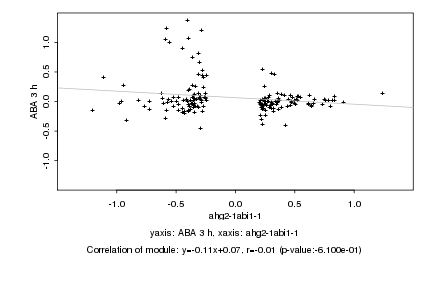
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ahg2-1abi1-1 |
Log2 signal ratio
ABA 3 h |
| 1 |
255471_at |
AT4G03050
|
AOP3 |
1.238 |
0.137 |
| 2 |
252345_at |
AT3G48640
|
unknown |
0.910 |
-0.001 |
| 3 |
253532_at |
AT4G31570
|
unknown |
0.832 |
0.018 |
| 4 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.831 |
0.098 |
| 5 |
244901_at |
ATMG00640
|
ORF25 |
0.811 |
0.024 |
| 6 |
254566_at |
AT4G19240
|
unknown |
0.796 |
-0.084 |
| 7 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.782 |
0.034 |
| 8 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.758 |
0.021 |
| 9 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.749 |
0.041 |
| 10 |
244945_at |
ATMG00110
|
ABCI2, ATP-binding cassette I2, CCB206, cytochrome C biogenesis 206 |
0.726 |
-0.035 |
| 11 |
257417_at |
AT1G10110
|
[F-box family protein] |
0.658 |
0.046 |
| 12 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.654 |
-0.023 |
| 13 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
0.635 |
-0.055 |
| 14 |
250135_at |
AT5G15360
|
unknown |
0.634 |
-0.073 |
| 15 |
248695_at |
AT5G48350
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.623 |
0.106 |
| 16 |
253689_at |
AT4G29770
|
unknown |
0.613 |
-0.019 |
| 17 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
0.607 |
-0.044 |
| 18 |
263515_at |
AT2G21640
|
unknown |
0.541 |
0.074 |
| 19 |
257057_at |
AT3G15310
|
unknown |
0.531 |
0.085 |
| 20 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.518 |
0.101 |
| 21 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.515 |
0.030 |
| 22 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.506 |
0.047 |
| 23 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
0.505 |
-0.049 |
| 24 |
244925_at |
ATMG00510
|
NAD7, NADH dehydrogenase subunit 7 |
0.496 |
0.034 |
| 25 |
248734_at |
AT5G48090
|
ELP1, EDM2-like protein1 |
0.490 |
-0.017 |
| 26 |
257321_at |
ATMG01130
|
ORF106F |
0.479 |
0.078 |
| 27 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.469 |
0.013 |
| 28 |
249461_at |
AT5G39640
|
[Putative endonuclease or glycosyl hydrolase] |
0.465 |
-0.012 |
| 29 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
0.460 |
0.118 |
| 30 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
0.460 |
-0.053 |
| 31 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
0.439 |
0.035 |
| 32 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.432 |
-0.074 |
| 33 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.421 |
-0.397 |
| 34 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.405 |
0.117 |
| 35 |
266729_at |
AT2G03130
|
[Ribosomal protein L12/ ATP-dependent Clp protease adaptor protein ClpS family protein] |
0.386 |
0.123 |
| 36 |
246888_at |
AT5G26270
|
unknown |
0.382 |
-0.100 |
| 37 |
260473_at |
AT1G10880
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.371 |
0.026 |
| 38 |
256188_at |
AT1G30160
|
unknown |
0.361 |
0.052 |
| 39 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
0.361 |
-0.120 |
| 40 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.358 |
0.021 |
| 41 |
248835_at |
AT5G47250
|
[LRR and NB-ARC domains-containing disease resistance protein] |
0.351 |
-0.009 |
| 42 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.348 |
0.136 |
| 43 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.346 |
0.013 |
| 44 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.338 |
-0.044 |
| 45 |
248394_at |
AT5G52070
|
[Agenet domain-containing protein] |
0.329 |
0.017 |
| 46 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.323 |
0.460 |
| 47 |
248148_at |
AT5G54930
|
[AT hook motif-containing protein] |
0.314 |
-0.154 |
| 48 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.313 |
-0.110 |
| 49 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.310 |
-0.019 |
| 50 |
245454_at |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.304 |
-0.046 |
| 51 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.303 |
-0.002 |
| 52 |
253945_at |
AT4G27050
|
[F-box/RNI-like superfamily protein] |
0.303 |
-0.116 |
| 53 |
255613_at |
AT4G01270
|
[RING/U-box superfamily protein] |
0.301 |
-0.048 |
| 54 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.298 |
0.488 |
| 55 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
0.289 |
-0.052 |
| 56 |
245446_at |
AT4G16800
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.284 |
-0.090 |
| 57 |
257323_at |
ATMG01200
|
ORF294 |
0.280 |
0.118 |
| 58 |
264830_at |
AT1G03710
|
[Cystatin/monellin superfamily protein] |
0.275 |
0.079 |
| 59 |
258741_at |
AT3G05790
|
LON4, lon protease 4 |
0.274 |
0.005 |
| 60 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.267 |
0.017 |
| 61 |
244921_s_at |
ATMG01000
|
ORF114 |
0.265 |
0.010 |
| 62 |
247250_at |
AT5G64630
|
FAS2, FASCIATA 2, MUB3.9, NFB01, NFB1 |
0.260 |
-0.066 |
| 63 |
247015_at |
AT5G66960
|
[Prolyl oligopeptidase family protein] |
0.260 |
-0.052 |
| 64 |
247345_at |
AT5G63760
|
ARI15, ARIADNE 15, ATARI15, ARABIDOPSIS ARIADNE 15 |
0.253 |
-0.042 |
| 65 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.251 |
-0.223 |
| 66 |
254287_at |
AT4G22960
|
unknown |
0.250 |
0.054 |
| 67 |
256830_at |
AT3G22860
|
ATEIF3C-2, ATTIF3C2, EIF3C-2, EUKARYOTIC INITIATION FACTOR 3C-2, TIF3C2, eukaryotic translation initiation factor 3 subunit C2 |
0.250 |
0.011 |
| 68 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
0.249 |
0.049 |
| 69 |
261221_at |
AT1G19960
|
unknown |
0.249 |
-0.145 |
| 70 |
258380_at |
AT3G16650
|
PRL2, Pleiotropic regulatory locus 2 |
0.246 |
-0.022 |
| 71 |
262421_at |
AT1G50290
|
unknown |
0.242 |
0.059 |
| 72 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.241 |
0.008 |
| 73 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.237 |
0.263 |
| 74 |
260464_at |
AT1G10920
|
LOV1, LOCUS ORCHESTRATING VICTORIN EFFECTS1 |
0.236 |
0.030 |
| 75 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.230 |
-0.102 |
| 76 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.226 |
-0.378 |
| 77 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.224 |
0.555 |
| 78 |
254867_at |
AT4G12240
|
[zinc finger (C2H2 type) family protein] |
0.223 |
-0.081 |
| 79 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.223 |
-0.132 |
| 80 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.222 |
-0.045 |
| 81 |
262508_at |
AT1G11300
|
[protein serine/threonine kinases] |
0.222 |
0.050 |
| 82 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.221 |
-0.059 |
| 83 |
251864_at |
AT3G54920
|
PMR6, powdery mildew resistant 6 |
0.219 |
-0.297 |
| 84 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.218 |
-0.062 |
| 85 |
248877_at |
AT5G46140
|
unknown |
0.218 |
-0.046 |
| 86 |
258198_at |
AT3G14020
|
NF-YA6, nuclear factor Y, subunit A6 |
0.217 |
-0.090 |
| 87 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
0.211 |
-0.035 |
| 88 |
249386_at |
AT5G40060
|
[Disease resistance protein (NBS-LRR class) family] |
0.209 |
0.022 |
| 89 |
252551_at |
AT3G45880
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.204 |
-0.032 |
| 90 |
265154_at |
AT1G30960
|
[GTP-binding family protein] |
0.203 |
-0.220 |
| 91 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.202 |
-0.003 |
| 92 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-1.207 |
-0.140 |
| 93 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-1.112 |
0.418 |
| 94 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.984 |
-0.021 |
| 95 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
-0.962 |
0.017 |
| 96 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
-0.945 |
0.281 |
| 97 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.919 |
-0.307 |
| 98 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.823 |
0.029 |
| 99 |
248016_at |
AT5G56380
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.768 |
-0.081 |
| 100 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.731 |
-0.119 |
| 101 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
-0.729 |
0.005 |
| 102 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
-0.627 |
0.140 |
| 103 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.621 |
0.056 |
| 104 |
249331_at |
AT5G40950
|
RPL27, ribosomal protein large subunit 27 |
-0.613 |
-0.021 |
| 105 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.596 |
-0.286 |
| 106 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
-0.595 |
1.054 |
| 107 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.585 |
1.247 |
| 108 |
248847_at |
AT5G46510
|
VICTL, VARIATION IN COMPOUND TRIGGERED ROOT growth response-like |
-0.581 |
-0.151 |
| 109 |
247988_at |
AT5G56910
|
[Proteinase inhibitor I25, cystatin, conserved region] |
-0.575 |
-0.010 |
| 110 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.572 |
0.035 |
| 111 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.560 |
1.014 |
| 112 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
-0.540 |
0.017 |
| 113 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
-0.530 |
0.072 |
| 114 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.522 |
-0.073 |
| 115 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
-0.500 |
0.005 |
| 116 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
-0.487 |
-0.140 |
| 117 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.486 |
-0.043 |
| 118 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.482 |
0.084 |
| 119 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
-0.450 |
-0.181 |
| 120 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.447 |
0.906 |
| 121 |
248954_at |
AT5G45420
|
maMYB, membrane anchored MYB |
-0.447 |
-0.116 |
| 122 |
254586_at |
AT4G19510
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-0.440 |
0.026 |
| 123 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.432 |
-0.160 |
| 124 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.430 |
-0.194 |
| 125 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.415 |
0.050 |
| 126 |
255121_at |
AT4G08480
|
MAPKKK9, mitogen-activated protein kinase kinase kinase 9, MEKK2, MAPK/ERK KINASE KINASE 2, SUMM1, suppressor of mkk1 mkk2 1 |
-0.412 |
0.006 |
| 127 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.405 |
1.387 |
| 128 |
249355_at |
AT5G40500
|
unknown |
-0.401 |
0.187 |
| 129 |
255627_at |
AT4G00955
|
unknown |
-0.400 |
-0.142 |
| 130 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.399 |
-0.048 |
| 131 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.397 |
1.067 |
| 132 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.396 |
-0.156 |
| 133 |
248976_at |
AT5G44930
|
ARAD2, ARABINAN DEFICIENT 2 |
-0.395 |
-0.081 |
| 134 |
256603_at |
AT3G28270
|
unknown |
-0.393 |
0.213 |
| 135 |
248722_at |
AT5G47810
|
PFK2, phosphofructokinase 2 |
-0.387 |
-0.083 |
| 136 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.384 |
-0.124 |
| 137 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.383 |
0.021 |
| 138 |
250778_at |
AT5G05500
|
MOP10 |
-0.378 |
-0.026 |
| 139 |
252937_at |
AT4G39180
|
ATSEC14, ARABIDOPSIS THALIANA SECRETION 14, SEC14, SECRETION 14 |
-0.366 |
0.072 |
| 140 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.366 |
-0.034 |
| 141 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.366 |
0.750 |
| 142 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.365 |
-0.033 |
| 143 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.362 |
0.287 |
| 144 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.361 |
-0.055 |
| 145 |
247167_at |
AT5G65850
|
[F-box and associated interaction domains-containing protein] |
-0.358 |
-0.098 |
| 146 |
247934_at |
AT5G57000
|
unknown |
-0.358 |
0.116 |
| 147 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.357 |
0.031 |
| 148 |
248977_at |
AT5G45020
|
[Glutathione S-transferase family protein] |
-0.351 |
-0.020 |
| 149 |
267191_at |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
-0.349 |
0.067 |
| 150 |
248079_at |
AT5G55790
|
unknown |
-0.349 |
-0.183 |
| 151 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.347 |
0.126 |
| 152 |
248706_at |
AT5G48530
|
unknown |
-0.342 |
-0.074 |
| 153 |
252822_at |
AT4G39955
|
[alpha/beta-Hydrolases superfamily protein] |
-0.337 |
0.268 |
| 154 |
245453_at |
AT4G16900
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.335 |
0.036 |
| 155 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
-0.320 |
-0.075 |
| 156 |
264459_at |
AT1G10160
|
[non-LTR retrotransposon family (LINE), has a 3.9e-39 P-value blast match to GB:AAA67727 reverse transcriptase (LINE-element) (Mus musculus)] |
-0.319 |
-0.089 |
| 157 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-0.318 |
0.821 |
| 158 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.317 |
0.462 |
| 159 |
249123_at |
AT5G43760
|
KCS20, 3-ketoacyl-CoA synthase 20 |
-0.316 |
0.151 |
| 160 |
247486_at |
AT5G62140
|
unknown |
-0.309 |
0.069 |
| 161 |
248293_at |
AT5G53050
|
[alpha/beta-Hydrolases superfamily protein] |
-0.306 |
0.067 |
| 162 |
252888_at |
AT4G39210
|
APL3 |
-0.303 |
0.672 |
| 163 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.299 |
-0.441 |
| 164 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.299 |
0.116 |
| 165 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.295 |
0.040 |
| 166 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.292 |
1.215 |
| 167 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.291 |
-0.007 |
| 168 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.290 |
0.019 |
| 169 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
-0.280 |
0.450 |
| 170 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.280 |
-0.160 |
| 171 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
-0.279 |
0.535 |
| 172 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.276 |
-0.069 |
| 173 |
254299_at |
AT4G22920
|
ATNYE1, NON-YELLOWING 1, NYE1, NON-YELLOWING 1, SGR, STAY-GREEN |
-0.275 |
0.410 |
| 174 |
262349_at |
AT2G48130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.271 |
0.254 |
| 175 |
249797_at |
AT5G23750
|
[Remorin family protein] |
-0.268 |
0.074 |
| 176 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.261 |
0.084 |
| 177 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
-0.259 |
0.056 |
| 178 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
-0.258 |
0.081 |
| 179 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
-0.254 |
0.137 |
| 180 |
245327_at |
AT4G16850
|
unknown |
-0.250 |
0.021 |
| 181 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
-0.246 |
0.455 |