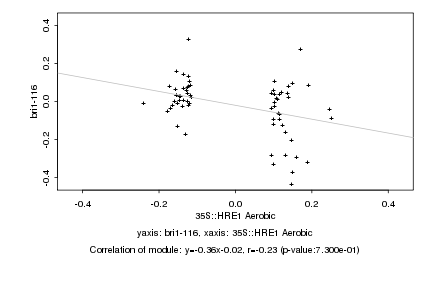
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S::HRE1 Aerobic |
Log2 signal ratio
bri1-116 |
| 1 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
0.250 |
-0.086 |
| 2 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.245 |
-0.039 |
| 3 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.189 |
0.086 |
| 4 |
258203_at |
AT3G13950
|
unknown |
0.188 |
-0.320 |
| 5 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.169 |
0.274 |
| 6 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.159 |
-0.290 |
| 7 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.148 |
-0.368 |
| 8 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.147 |
0.098 |
| 9 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
0.146 |
-0.203 |
| 10 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.146 |
-0.431 |
| 11 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
0.138 |
0.026 |
| 12 |
265496_at |
AT2G15700
|
[copia-like retrotransposon family, has a 2.4e-308 P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.137 |
0.081 |
| 13 |
260422_at |
AT1G69630
|
[F-box/RNI-like superfamily protein] |
0.135 |
0.044 |
| 14 |
257323_at |
ATMG01200
|
ORF294 |
0.130 |
-0.280 |
| 15 |
266827_at |
AT2G22920
|
SCPL12, serine carboxypeptidase-like 12 |
0.129 |
-0.159 |
| 16 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.122 |
-0.121 |
| 17 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.119 |
0.050 |
| 18 |
250585_at |
AT5G07620
|
[Protein kinase superfamily protein] |
0.115 |
-0.065 |
| 19 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.115 |
-0.093 |
| 20 |
263851_at |
AT2G04460
|
unknown |
0.114 |
0.041 |
| 21 |
248535_at |
AT5G50120
|
[Transducin/WD40 repeat-like superfamily protein] |
0.112 |
-0.062 |
| 22 |
245345_at |
AT4G16640
|
[Matrixin family protein] |
0.108 |
0.014 |
| 23 |
249362_at |
AT5G40550
|
AtSGF29b, SGF29b, SaGa associated Factor 29 b |
0.107 |
0.018 |
| 24 |
262255_at |
AT1G53790
|
[F-box and associated interaction domains-containing protein] |
0.102 |
-0.022 |
| 25 |
254816_at |
AT4G12440
|
APT4, adenine phosphoribosyl transferase 4 |
0.100 |
0.110 |
| 26 |
247797_at |
AT5G58780
|
AtcPT5, AtHEPS, cPT5, cis -prenyltransferase 5, HEPS, heptaprenyl diphosphate synthase |
0.100 |
0.041 |
| 27 |
256131_at |
AT1G13600
|
AtbZIP58, basic leucine-zipper 58, bZIP58, basic leucine-zipper 58 |
0.100 |
-0.003 |
| 28 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.098 |
-0.117 |
| 29 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
0.098 |
-0.090 |
| 30 |
246262_at |
AT1G31790
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.097 |
0.058 |
| 31 |
249009_at |
AT5G44610
|
MAP18, microtubule-associated protein 18, PCAP2, PLASMA MEMBRANE ASSOCIATED CA2+-BINDING PROTEIN-2 |
0.097 |
-0.326 |
| 32 |
259827_at |
AT1G72270
|
unknown |
0.094 |
0.043 |
| 33 |
265907_at |
AT2G25650
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
0.094 |
-0.035 |
| 34 |
266640_at |
AT2G35585
|
unknown |
0.092 |
-0.283 |
| 35 |
248367_at |
AT5G52360
|
ADF10, actin depolymerizing factor 10 |
-0.242 |
-0.010 |
| 36 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
-0.179 |
-0.052 |
| 37 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.173 |
0.084 |
| 38 |
267585_s_at |
AT2G42090
|
ACT9, actin 9 |
-0.172 |
-0.032 |
| 39 |
264601_at |
AT1G04540
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.167 |
-0.020 |
| 40 |
265052_at |
AT1G51990
|
[O-methyltransferase family protein] |
-0.161 |
0.002 |
| 41 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.159 |
0.066 |
| 42 |
257629_at |
AT3G26140
|
[Cellulase (glycosyl hydrolase family 5) protein] |
-0.156 |
0.035 |
| 43 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
-0.155 |
0.033 |
| 44 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.155 |
0.162 |
| 45 |
247648_at |
AT5G60020
|
ATLAC17, LAC17, laccase 17 |
-0.153 |
-0.129 |
| 46 |
262998_at |
AT1G54280
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.153 |
-0.010 |
| 47 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.148 |
0.007 |
| 48 |
256096_at |
AT1G13650
|
unknown |
-0.148 |
0.029 |
| 49 |
261998_at |
AT1G33813
|
[copia-like retrotransposon family, has a 5.8e-39 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.145 |
0.028 |
| 50 |
246937_at |
AT5G25370
|
PLDALPHA3, phospholipase D alpha 3 |
-0.140 |
-0.021 |
| 51 |
263794_at |
AT2G24620
|
[S-locus glycoprotein family protein] |
-0.138 |
0.147 |
| 52 |
264194_at |
AT1G22720
|
[Protein kinase superfamily protein] |
-0.137 |
0.008 |
| 53 |
266263_at |
AT2G27570
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.136 |
0.071 |
| 54 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
-0.131 |
-0.171 |
| 55 |
250889_at |
AT5G04500
|
[glycosyltransferase family protein 47] |
-0.129 |
0.059 |
| 56 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.128 |
0.076 |
| 57 |
260019_at |
AT1G41825
|
[gypsy-like retrotransposon family (Athila), has a 1.9e-98 P-value blast match to gb|AAL06422.1|AF378081_1 reverse transcriptase (Athila4) (Arabidopsis thaliana) (Gypsy_Ty3-family)] |
-0.128 |
0.046 |
| 58 |
260562_at |
AT2G43850
|
[Integrin-linked protein kinase family] |
-0.126 |
0.005 |
| 59 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.124 |
0.327 |
| 60 |
249830_at |
AT5G23300
|
PYRD, pyrimidine d |
-0.124 |
-0.017 |
| 61 |
247772_at |
AT5G58610
|
[PHD finger transcription factor, putative] |
-0.124 |
0.080 |
| 62 |
263290_at |
AT2G10930
|
unknown |
-0.123 |
0.136 |
| 63 |
262937_at |
AT1G79560
|
EMB1047, EMBRYO DEFECTIVE 1047, EMB156, EMBRYO DEFECTIVE 156, EMB36, EMBRYO DEFECTIVE 36, FTSH12, FTSH protease 12 |
-0.121 |
0.110 |
| 64 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
-0.121 |
-0.007 |
| 65 |
255124_at |
AT4G08560
|
APUM15, pumilio 15, PUM15, pumilio 15 |
-0.120 |
0.035 |
| 66 |
262266_at |
AT1G42450
|
[Mutator-like transposase family, has a 4.3e-96 P-value blast match to Q9SHN7 /450-633 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.118 |
0.085 |
| 67 |
258305_at |
AT3G30650
|
unknown |
-0.117 |
0.025 |