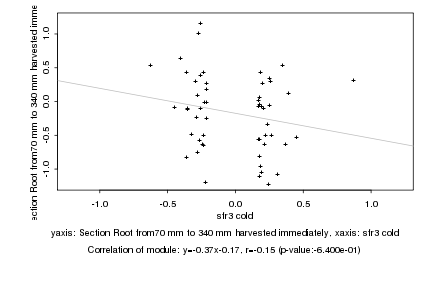
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
sfr3 cold |
Log2 signal ratio
Section Root from70 mm to 340 mm harvested immediately |
| 1 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.866 |
0.320 |
| 2 |
245553_at |
AT4G15370
|
BARS1, baruol synthase 1, PEN2, PENTACYCLIC TRITERPENE SYNTHASE 2 |
0.443 |
-0.530 |
| 3 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
0.384 |
0.132 |
| 4 |
262510_at |
AT1G11270
|
[F-box and associated interaction domains-containing protein] |
0.363 |
-0.629 |
| 5 |
256966_at |
AT3G13400
|
sks13, SKU5 similar 13 |
0.345 |
0.540 |
| 6 |
248368_at |
AT5G51950
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.306 |
-1.079 |
| 7 |
260886_at |
AT1G29200
|
[O-fucosyltransferase family protein] |
0.265 |
-0.498 |
| 8 |
257229_at |
AT3G16490
|
IQD26, IQ-domain 26 |
0.254 |
0.310 |
| 9 |
267160_at |
AT2G37670
|
[Transducin/WD40 repeat-like superfamily protein] |
0.250 |
-0.056 |
| 10 |
247633_at |
AT5G60470
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.245 |
0.347 |
| 11 |
251180_at |
AT3G62640
|
unknown |
0.238 |
-1.214 |
| 12 |
245614_at |
AT4G14460
|
[copia-like retrotransposon family, has a 4.2e-253 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.233 |
-0.338 |
| 13 |
263018_at |
AT2G17610
|
[non-LTR retrotransposon family (LINE), has a 3.6e-21 P-value blast match to GB:AAA39398 ORF2 (Mus musculus) (LINE-element)] |
0.219 |
-0.496 |
| 14 |
249704_at |
AT5G35550
|
ATMYB123, MYB DOMAIN PROTEIN 123, ATTT2, MYB123, MYB DOMAIN PROTEIN 123, TT2, TRANSPARENT TESTA 2 |
0.210 |
-0.631 |
| 15 |
250514_at |
AT5G09550
|
GDI, RAB GDP-dissociation inhibitor |
0.205 |
-0.091 |
| 16 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.193 |
0.273 |
| 17 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
0.187 |
-0.070 |
| 18 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
0.186 |
-1.049 |
| 19 |
263991_at |
AT2G13020
|
[gypsy-like retrotransposon family (Athila), has a 7.3e-101 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.180 |
-0.958 |
| 20 |
256893_x_at |
AT3G30660
|
unknown |
0.178 |
0.444 |
| 21 |
267206_at |
AT2G30830
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.177 |
-0.811 |
| 22 |
267624_at |
AT2G39660
|
BIK1, botrytis-induced kinase1 |
0.177 |
-0.030 |
| 23 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
0.174 |
-1.108 |
| 24 |
265127_at |
AT1G55560
|
sks14, SKU5 similar 14 |
0.171 |
0.062 |
| 25 |
246592_at |
AT5G14890
|
[NHL domain-containing protein] |
0.170 |
-0.559 |
| 26 |
257737_at |
AT3G27440
|
UKL5, uridine kinase-like 5 |
0.169 |
-0.551 |
| 27 |
249882_at |
AT5G22890
|
STOP2, sensitive to proton rhizotoxicity 2 |
0.169 |
0.027 |
| 28 |
253193_at |
AT4G35380
|
[SEC7-like guanine nucleotide exchange family protein] |
0.169 |
-0.071 |
| 29 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.630 |
0.546 |
| 30 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.450 |
-0.076 |
| 31 |
250788_at |
AT5G05570
|
[transducin family protein / WD-40 repeat family protein] |
-0.409 |
0.637 |
| 32 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
-0.361 |
0.444 |
| 33 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
-0.361 |
-0.822 |
| 34 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.359 |
-0.093 |
| 35 |
248533_at |
AT5G50020
|
[DHHC-type zinc finger family protein] |
-0.355 |
-0.115 |
| 36 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
-0.324 |
-0.476 |
| 37 |
246115_at |
AT5G20300
|
Toc90, translocon at the outer envelope membrane of chloroplasts 90 |
-0.298 |
0.311 |
| 38 |
262538_at |
AT1G17140
|
ICR1, interactor of constitutive active rops 1, RIP1, ROP INTERACTIVE PARTNER 1 |
-0.294 |
-0.229 |
| 39 |
262596_at |
AT1G15080
|
ATLPP2, LIPID PHOSPHATE PHOSPHATASE 2, ATPAP2, PHOSPHATIDIC ACID PHOSPHATASE 2, LPP2, lipid phosphate phosphatase 2 |
-0.281 |
-0.754 |
| 40 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.280 |
0.098 |
| 41 |
257875_at |
AT3G17120
|
unknown |
-0.274 |
1.016 |
| 42 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
-0.268 |
-0.573 |
| 43 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.261 |
-0.103 |
| 44 |
265522_at |
AT2G06210
|
ELF8, EARLY FLOWERING 8, VIP6, VERNALIZATION INDEPENDENCE 6 |
-0.260 |
1.168 |
| 45 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.258 |
0.391 |
| 46 |
267036_at |
AT2G38465
|
unknown |
-0.243 |
-0.629 |
| 47 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.241 |
-0.492 |
| 48 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.238 |
-0.643 |
| 49 |
249943_at |
AT5G22280
|
unknown |
-0.237 |
0.433 |
| 50 |
252001_at |
AT3G52750
|
FTSZ2-2 |
-0.229 |
-0.010 |
| 51 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.221 |
-1.196 |
| 52 |
264340_at |
AT1G70280
|
[NHL domain-containing protein] |
-0.220 |
-0.000 |
| 53 |
253292_at |
AT4G33985
|
unknown |
-0.219 |
0.182 |
| 54 |
248485_at |
AT5G51050
|
APC2, ATP/phosphate carrier 2 |
-0.217 |
0.277 |
| 55 |
250936_at |
AT5G03120
|
unknown |
-0.217 |
-0.244 |