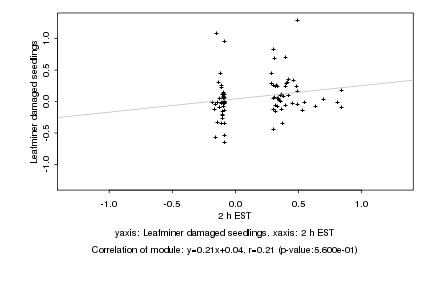
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
2 h EST |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
247579_at |
AT5G61320
|
CYP89A3, cytochrome P450, family 89, subfamily A, polypeptide 3 |
0.838 |
-0.086 |
| 2 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.836 |
0.186 |
| 3 |
261243_at |
AT1G20180
|
unknown |
0.804 |
-0.009 |
| 4 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.692 |
0.043 |
| 5 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
0.631 |
-0.073 |
| 6 |
258960_at |
AT3G10590
|
[Duplicated homeodomain-like superfamily protein] |
0.541 |
-0.011 |
| 7 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
0.528 |
-0.141 |
| 8 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.491 |
-0.037 |
| 9 |
245082_at |
AT2G23270
|
unknown |
0.484 |
0.172 |
| 10 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.484 |
1.304 |
| 11 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
0.479 |
0.243 |
| 12 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.454 |
0.339 |
| 13 |
250965_at |
AT5G03020
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.451 |
-0.023 |
| 14 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.420 |
0.351 |
| 15 |
246193_at |
AT5G20880
|
[retrotransposon family] |
0.417 |
0.108 |
| 16 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
0.409 |
0.314 |
| 17 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.400 |
0.298 |
| 18 |
253124_at |
AT4G36030
|
ARO3, armadillo repeat only 3 |
0.395 |
0.240 |
| 19 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.395 |
0.701 |
| 20 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.393 |
-0.062 |
| 21 |
265582_at |
AT2G20030
|
[RING/U-box superfamily protein] |
0.373 |
0.080 |
| 22 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
0.369 |
-0.337 |
| 23 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
0.364 |
0.124 |
| 24 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.360 |
-0.120 |
| 25 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.356 |
0.105 |
| 26 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.351 |
0.015 |
| 27 |
248038_at |
AT5G55980
|
[serine-rich protein-related] |
0.346 |
0.029 |
| 28 |
264657_at |
AT1G09100
|
RPT5B, 26S proteasome AAA-ATPase subunit RPT5B |
0.336 |
0.059 |
| 29 |
247003_at |
AT5G67550
|
unknown |
0.331 |
-0.070 |
| 30 |
265703_at |
AT2G03430
|
[Ankyrin repeat family protein] |
0.329 |
0.061 |
| 31 |
250821_at |
AT5G05190
|
unknown |
0.328 |
0.247 |
| 32 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
0.320 |
0.267 |
| 33 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.316 |
0.254 |
| 34 |
250849_at |
AT5G04410
|
anac078, Arabidopsis NAC domain containing protein 78, NAC2, NAC domain containing protein 2 |
0.314 |
-0.050 |
| 35 |
254933_at |
AT4G11130
|
RDR2, RNA-dependent RNA polymerase 2, SMD1, SILENCING MOVEMENT DEFICIENT 1 |
0.312 |
-0.158 |
| 36 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.309 |
0.690 |
| 37 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.308 |
0.069 |
| 38 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
0.297 |
0.063 |
| 39 |
265880_at |
AT2G42300
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.296 |
-0.444 |
| 40 |
255647_at |
AT4G00900
|
ATECA2, ARABIDOPSIS THALIANA ER-TYPE CA2+-ATPASE 2, ECA2, ER-type Ca2+-ATPase 2 |
0.296 |
-0.118 |
| 41 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.296 |
0.840 |
| 42 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
0.294 |
0.259 |
| 43 |
248820_at |
AT5G47060
|
unknown |
0.282 |
0.295 |
| 44 |
260666_at |
AT1G19300
|
ATGATL1, GATL1, GALACTURONOSYLTRANSFERASE-LIKE 1, GLZ1, GAOLAOZHUANGREN 1, PARVUS, PARVUS |
0.278 |
0.449 |
| 45 |
264296_at |
AT1G78720
|
[SecY protein transport family protein] |
-0.184 |
-0.012 |
| 46 |
266257_at |
AT2G27820
|
ADT3, arogenate dehydratase 3, PD1, prephenate dehydratase 1 |
-0.171 |
-0.125 |
| 47 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.165 |
-0.559 |
| 48 |
254658_at |
AT4G18230
|
unknown |
-0.162 |
-0.034 |
| 49 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-0.157 |
1.091 |
| 50 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
-0.145 |
-0.323 |
| 51 |
261971_at |
AT1G65990
|
[type 2 peroxiredoxin-related / thiol specific antioxidant / mal allergen family protein] |
-0.143 |
-0.008 |
| 52 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
-0.135 |
0.311 |
| 53 |
267286_at |
AT2G23640
|
RTNLB13, Reticulan like protein B13 |
-0.133 |
0.059 |
| 54 |
264775_at |
AT1G22880
|
ATCEL5, ARABIDOPSIS THALIANA CELLULASE 5, ATGH9B4, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B4, CEL5, cellulase 5 |
-0.130 |
-0.083 |
| 55 |
256959_at |
AT3G13440
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.128 |
-0.088 |
| 56 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
-0.126 |
0.459 |
| 57 |
258577_at |
AT3G04220
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.114 |
0.264 |
| 58 |
267625_at |
AT2G42240
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.114 |
-0.028 |
| 59 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
-0.113 |
0.238 |
| 60 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.111 |
-0.346 |
| 61 |
247253_at |
AT5G64790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.111 |
-0.006 |
| 62 |
260790_at |
AT1G06240
|
unknown |
-0.110 |
-0.265 |
| 63 |
250566_at |
AT5G08070
|
TCP17, TCP domain protein 17 |
-0.108 |
-0.206 |
| 64 |
256446_at |
AT3G11110
|
[RING/U-box superfamily protein] |
-0.106 |
-0.210 |
| 65 |
251728_at |
AT3G56300
|
[Cysteinyl-tRNA synthetase, class Ia family protein] |
-0.104 |
-0.151 |
| 66 |
246537_at |
AT5G15940
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.104 |
0.112 |
| 67 |
265057_at |
AT1G52140
|
unknown |
-0.102 |
0.069 |
| 68 |
255385_at |
AT4G03610
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.102 |
0.049 |
| 69 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
-0.101 |
0.142 |
| 70 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.097 |
0.155 |
| 71 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
-0.096 |
-0.027 |
| 72 |
249747_at |
AT5G24600
|
unknown |
-0.096 |
0.085 |
| 73 |
247762_at |
AT5G59170
|
[Proline-rich extensin-like family protein] |
-0.095 |
-0.069 |
| 74 |
245103_at |
AT2G41590
|
unknown |
-0.095 |
0.049 |
| 75 |
245057_at |
AT2G26490
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.095 |
-0.003 |
| 76 |
264909_at |
AT2G17300
|
unknown |
-0.094 |
-0.537 |
| 77 |
263013_at |
AT1G23380
|
KNAT6, KNOTTED1-like homeobox gene 6, KNAT6L, KNAT6S |
-0.094 |
-0.021 |
| 78 |
254145_at |
AT4G24700
|
unknown |
-0.091 |
-0.645 |
| 79 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
-0.091 |
0.964 |
| 80 |
248416_at |
AT5G51630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.091 |
-0.128 |
| 81 |
246534_at |
AT5G15890
|
TBL21, TRICHOME BIREFRINGENCE-LIKE 21 |
-0.091 |
-0.015 |
| 82 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
-0.091 |
0.066 |
| 83 |
255358_at |
AT4G03940
|
unknown |
-0.090 |
-0.013 |
| 84 |
266390_at |
AT2G32310
|
[CCT motif family protein] |
-0.089 |
0.010 |
| 85 |
266384_at |
AT2G14660
|
unknown |
-0.088 |
-0.342 |
| 86 |
254708_at |
AT4G17895
|
UBP20, ubiquitin-specific protease 20 |
-0.087 |
0.126 |
| 87 |
253699_at |
AT4G29800
|
PLA IVD, PLP8, PATATIN-like protein 8 |
-0.087 |
0.005 |