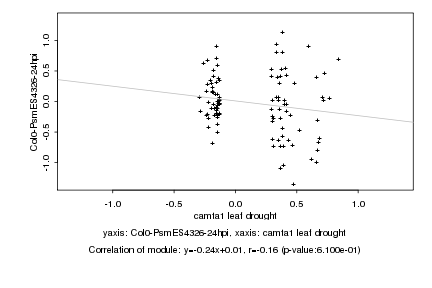
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
camta1 leaf drought |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.833 |
0.697 |
| 2 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.761 |
0.059 |
| 3 |
247324_at |
AT5G64190
|
unknown |
0.723 |
0.471 |
| 4 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
0.717 |
0.022 |
| 5 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.706 |
0.071 |
| 6 |
247474_at |
AT5G62280
|
unknown |
0.684 |
-0.600 |
| 7 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
0.674 |
-0.660 |
| 8 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
0.662 |
-0.799 |
| 9 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.661 |
-0.297 |
| 10 |
256569_at |
AT3G19550
|
unknown |
0.657 |
0.398 |
| 11 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
0.657 |
-0.994 |
| 12 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.619 |
-0.937 |
| 13 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.592 |
0.915 |
| 14 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.520 |
-0.459 |
| 15 |
260522_x_at |
AT2G41730
|
unknown |
0.474 |
0.309 |
| 16 |
256096_at |
AT1G13650
|
unknown |
0.473 |
-1.342 |
| 17 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.458 |
-0.710 |
| 18 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.447 |
-0.216 |
| 19 |
260081_at |
AT1G78170
|
unknown |
0.428 |
-0.628 |
| 20 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.414 |
-0.043 |
| 21 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.414 |
0.438 |
| 22 |
247882_at |
AT5G57785
|
unknown |
0.412 |
-0.148 |
| 23 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.401 |
0.547 |
| 24 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.397 |
-0.033 |
| 25 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.396 |
0.018 |
| 26 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.390 |
-0.735 |
| 27 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.384 |
-1.046 |
| 28 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.382 |
-0.433 |
| 29 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.382 |
0.811 |
| 30 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
0.376 |
1.139 |
| 31 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.376 |
-0.566 |
| 32 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.369 |
0.532 |
| 33 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.367 |
0.410 |
| 34 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
0.366 |
-0.729 |
| 35 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
0.364 |
-0.275 |
| 36 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
0.360 |
-1.080 |
| 37 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.356 |
0.311 |
| 38 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.352 |
-0.129 |
| 39 |
247149_at |
AT5G65660
|
[hydroxyproline-rich glycoprotein family protein] |
0.347 |
0.080 |
| 40 |
245383_at |
AT4G17810
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.347 |
-0.636 |
| 41 |
267193_at |
AT2G30900
|
TBL43, TRICHOME BIREFRINGENCE-LIKE 43 |
0.346 |
0.022 |
| 42 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.338 |
0.405 |
| 43 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.334 |
0.949 |
| 44 |
248302_at |
AT5G53160
|
PYL8, PYR1-like 8, RCAR3, regulatory components of ABA receptor 3 |
0.333 |
0.080 |
| 45 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.331 |
0.813 |
| 46 |
251080_at |
AT5G02010
|
ATROPGEF7, ROPGEF7, ROP (rho of plants) guanine nucleotide exchange factor 7 |
0.308 |
-0.268 |
| 47 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
0.307 |
-0.725 |
| 48 |
259373_at |
AT1G69160
|
unknown |
0.301 |
-0.617 |
| 49 |
246952_at |
AT5G04820
|
ATOFP13, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 13, OFP13, ovate family protein 13 |
0.298 |
-0.239 |
| 50 |
257543_at |
AT3G28960
|
[Transmembrane amino acid transporter family protein] |
0.297 |
-0.316 |
| 51 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
0.296 |
0.029 |
| 52 |
251427_at |
AT3G60130
|
BGLU16, beta glucosidase 16 |
0.293 |
0.540 |
| 53 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.292 |
0.410 |
| 54 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
0.292 |
-0.119 |
| 55 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
-0.294 |
0.069 |
| 56 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
-0.285 |
-0.157 |
| 57 |
259043_at |
AT3G03440
|
[ARM repeat superfamily protein] |
-0.264 |
0.637 |
| 58 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.241 |
-0.218 |
| 59 |
258603_at |
AT3G02990
|
ATHSFA1E, heat shock transcription factor A1E, HSFA1E, heat shock transcription factor A1E |
-0.239 |
0.178 |
| 60 |
247197_at |
AT5G65240
|
[Leucine-rich repeat protein kinase family protein] |
-0.233 |
0.284 |
| 61 |
245961_at |
AT5G19670
|
[Exostosin family protein] |
-0.230 |
-0.205 |
| 62 |
259534_at |
AT1G12290
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.229 |
0.672 |
| 63 |
253078_at |
AT4G36180
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.224 |
-0.001 |
| 64 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.222 |
-0.415 |
| 65 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
-0.222 |
-0.265 |
| 66 |
250211_at |
AT5G13880
|
unknown |
-0.207 |
0.346 |
| 67 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
-0.198 |
0.306 |
| 68 |
265279_at |
AT2G28460
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.197 |
-0.102 |
| 69 |
256210_at |
AT1G50950
|
[INVOLVED IN: cell redox homeostasis] |
-0.195 |
0.150 |
| 70 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.189 |
-0.687 |
| 71 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.188 |
0.169 |
| 72 |
263868_at |
AT2G36840
|
ACR10, ACT domain repeats 10 |
-0.187 |
0.235 |
| 73 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.185 |
-0.039 |
| 74 |
258807_at |
AT3G04030
|
MYR2 |
-0.184 |
0.413 |
| 75 |
255782_at |
AT1G19850
|
ARF5, AUXIN RESPONSE FACTOR 5, IAA24, indole-3-acetic acid inducible 24, MP, MONOPTEROS |
-0.180 |
0.520 |
| 76 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.180 |
0.154 |
| 77 |
254933_at |
AT4G11130
|
RDR2, RNA-dependent RNA polymerase 2, SMD1, SILENCING MOVEMENT DEFICIENT 1 |
-0.172 |
-0.106 |
| 78 |
250685_at |
AT5G06670
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.171 |
-0.216 |
| 79 |
264535_at |
AT1G55690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.166 |
0.131 |
| 80 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
-0.160 |
-0.187 |
| 81 |
253854_at |
AT4G27900
|
[CCT motif family protein] |
-0.158 |
-0.138 |
| 82 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
-0.157 |
0.325 |
| 83 |
267177_at |
AT2G37580
|
[RING/U-box superfamily protein] |
-0.156 |
0.708 |
| 84 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
-0.155 |
0.916 |
| 85 |
267210_at |
AT2G30920
|
ATCOQ3, coenzyme Q 3, COQ3, COQ3, coenzyme Q 3, EMB3002, embryo defective 3002 |
-0.155 |
-0.090 |
| 86 |
257779_at |
AT3G26940
|
CDG1, CONSTITUTIVE DIFFERENTIAL GROWTH 1 |
-0.154 |
0.006 |
| 87 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.154 |
-0.494 |
| 88 |
261325_at |
AT1G44780
|
unknown |
-0.153 |
-0.217 |
| 89 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.152 |
-0.105 |
| 90 |
254186_at |
AT4G24010
|
ATCSLG1, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G1, CSLG1, cellulose synthase like G1 |
-0.151 |
0.131 |
| 91 |
264793_at |
AT1G08660
|
MGP2, MALE GAMETOPHYTE DEFECTIVE 2 |
-0.150 |
-0.360 |
| 92 |
266273_at |
AT2G29410
|
ATMTPB1, MTPB1, metal tolerance protein B1 |
-0.150 |
0.605 |
| 93 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
-0.149 |
-0.206 |
| 94 |
257112_at |
AT3G20120
|
CYP705A21, cytochrome P450, family 705, subfamily A, polypeptide 21 |
-0.148 |
-0.053 |
| 95 |
251383_at |
AT3G60740
|
CHO, CHAMPIGNON, EMB133, EMBRYO DEFECTIVE 133, TFC D, TUBULIN FOLDING COFACTOR D, TTN1, TITAN 1 |
-0.148 |
-0.256 |
| 96 |
251530_at |
AT3G58520
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.143 |
-0.032 |
| 97 |
256803_at |
AT3G20960
|
CYP705A33, cytochrome P450, family 705, subfamily A, polypeptide 33 |
-0.140 |
0.382 |
| 98 |
247231_at |
AT5G65230
|
AtMYB53, myb domain protein 53, MYB53, myb domain protein 53 |
-0.139 |
0.016 |
| 99 |
247105_at |
AT5G66630
|
DAR5, DA1-related protein 5 |
-0.137 |
0.345 |
| 100 |
245129_at |
AT2G45350
|
CRR4, CHLORORESPIRATORY REDUCTION 4 |
-0.137 |
-0.020 |
| 101 |
264874_at |
AT1G24240
|
[Ribosomal protein L19 family protein] |
-0.134 |
-0.031 |
| 102 |
264775_at |
AT1G22880
|
ATCEL5, ARABIDOPSIS THALIANA CELLULASE 5, ATGH9B4, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B4, CEL5, cellulase 5 |
-0.133 |
0.035 |
| 103 |
259641_at |
AT1G69020
|
[Prolyl oligopeptidase family protein] |
-0.131 |
-0.042 |
| 104 |
254800_at |
AT4G13070
|
[RNA-binding CRS1 / YhbY (CRM) domain protein] |
-0.131 |
0.022 |
| 105 |
245754_at |
AT1G35183
|
unknown |
-0.131 |
0.080 |
| 106 |
256899_at |
AT3G24660
|
TMKL1, transmembrane kinase-like 1 |
-0.131 |
-0.184 |
| 107 |
245575_at |
AT4G14760
|
NET1B, Networked 1B |
-0.130 |
-0.199 |