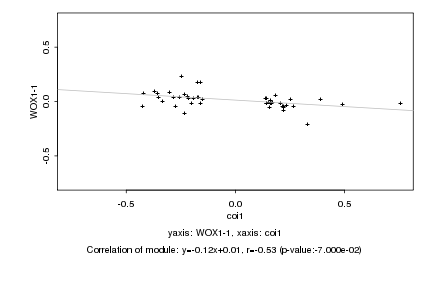
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
coi1 |
Log2 signal ratio
WOX1-1 |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.754 |
-0.016 |
| 2 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
0.490 |
-0.027 |
| 3 |
252314_at |
AT3G49400
|
[Transducin/WD40 repeat-like superfamily protein] |
0.385 |
0.021 |
| 4 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.327 |
-0.206 |
| 5 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.264 |
-0.040 |
| 6 |
246212_at |
AT4G36930
|
SPT, SPATULA |
0.251 |
0.027 |
| 7 |
262167_at |
AT1G74990
|
[RING/U-box superfamily protein] |
0.233 |
-0.031 |
| 8 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.219 |
-0.038 |
| 9 |
266275_at |
AT2G29370
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.219 |
-0.075 |
| 10 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.218 |
-0.047 |
| 11 |
253387_at |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
0.212 |
-0.041 |
| 12 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.206 |
-0.012 |
| 13 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.179 |
0.064 |
| 14 |
247422_at |
AT5G62810
|
ATPEX14, PED2, PEROXISOME DEFECTIVE 2, PEX14, peroxin 14 |
0.166 |
-0.008 |
| 15 |
250432_at |
AT5G10420
|
[MATE efflux family protein] |
0.163 |
-0.012 |
| 16 |
247659_at |
AT5G60040
|
NRPC1, nuclear RNA polymerase C1 |
0.160 |
0.018 |
| 17 |
252996_s_at |
AT4G38460
|
AtSSU, GGR, geranylgeranyl reductase, SSU, small subunit of Heterodimeric geranyl(geranyl)diphosphate synthase |
0.157 |
-0.014 |
| 18 |
253518_at |
AT4G31400
|
AtCTF7, CTF7, CHROMOSOME TRANSMISSION FIDELITY 7, ECO1 |
0.156 |
-0.018 |
| 19 |
259970_at |
AT1G76570
|
AtLHCB7, LHCB7, light-harvesting complex B7 |
0.155 |
-0.049 |
| 20 |
264641_at |
AT1G09130
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.151 |
-0.006 |
| 21 |
260660_at |
AT1G19485
|
[Transducin/WD40 repeat-like superfamily protein] |
0.140 |
-0.016 |
| 22 |
252333_at |
AT3G48830
|
[polynucleotide adenylyltransferase family protein / RNA recognition motif (RRM)-containing protein] |
0.140 |
-0.011 |
| 23 |
256700_at |
AT3G52260
|
[Pseudouridine synthase family protein] |
0.138 |
0.028 |
| 24 |
262588_at |
AT1G15130
|
[Endosomal targeting BRO1-like domain-containing protein] |
0.136 |
0.032 |
| 25 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
-0.426 |
-0.040 |
| 26 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
-0.424 |
0.077 |
| 27 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.371 |
0.094 |
| 28 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
-0.357 |
0.077 |
| 29 |
260395_at |
AT1G69780
|
ATHB13 |
-0.354 |
0.037 |
| 30 |
262913_at |
AT1G59960
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.337 |
0.006 |
| 31 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
-0.305 |
0.086 |
| 32 |
250738_at |
AT5G05730
|
AMT1, A-METHYL TRYPTOPHAN RESISTANT 1, ASA1, anthranilate synthase alpha subunit 1, JDL1, JASMONATE-INDUCED DEFECTIVE LATERAL ROOT 1, TRP5, TRYPTOPHAN BIOSYNTHESIS 5, WEI2, WEAK ETHYLENE INSENSITIVE 2 |
-0.284 |
0.037 |
| 33 |
257666_at |
AT3G20270
|
[lipid-binding serum glycoprotein family protein] |
-0.279 |
-0.043 |
| 34 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
-0.258 |
0.038 |
| 35 |
250875_at |
AT5G04020
|
[calmodulin binding] |
-0.251 |
0.239 |
| 36 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
-0.237 |
0.068 |
| 37 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.235 |
-0.102 |
| 38 |
250079_at |
AT5G16650
|
[Chaperone DnaJ-domain superfamily protein] |
-0.224 |
0.047 |
| 39 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.216 |
0.034 |
| 40 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
-0.203 |
-0.017 |
| 41 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.195 |
0.029 |
| 42 |
259385_at |
AT1G13470
|
unknown |
-0.178 |
0.044 |
| 43 |
261039_at |
AT1G17455
|
ELF4-L4, ELF4-like 4 |
-0.175 |
0.176 |
| 44 |
267077_at |
AT2G40970
|
MYBC1 |
-0.170 |
0.046 |
| 45 |
249025_at |
AT5G44720
|
[Molybdenum cofactor sulfurase family protein] |
-0.163 |
-0.013 |
| 46 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
-0.160 |
0.184 |
| 47 |
245330_at |
AT4G14930
|
[Survival protein SurE-like phosphatase/nucleotidase] |
-0.154 |
0.022 |