|
probeID |
AGICode |
Annotation |
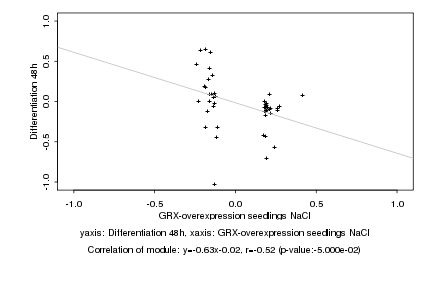
Log2 signal ratio
GRX-overexpression seedlings NaCl |
Log2 signal ratio
Differentiation 48h |
| 1 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.409 |
0.075 |
| 2 |
245068_at |
AT2G23260
|
UGT84B1, UDP-glucosyl transferase 84B1 |
0.268 |
-0.056 |
| 3 |
264966_at |
AT1G60570
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.260 |
-0.084 |
| 4 |
264297_at |
AT1G78710
|
TBL42, TRICHOME BIREFRINGENCE-LIKE 42 |
0.257 |
-0.100 |
| 5 |
262010_at |
AT1G35612
|
[expressed protein] |
0.241 |
-0.562 |
| 6 |
253634_at |
AT4G30590
|
AtENODL12, ENODL12, early nodulin-like protein 12 |
0.216 |
-0.140 |
| 7 |
250507_at |
AT5G09940
|
unknown |
0.212 |
-0.082 |
| 8 |
245823_at |
AT1G57906
|
unknown |
0.208 |
-0.098 |
| 9 |
264558_at |
AT1G09600
|
[Protein kinase superfamily protein] |
0.206 |
0.093 |
| 10 |
258591_at |
AT3G04360
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.198 |
-0.065 |
| 11 |
264505_at |
AT1G09380
|
UMAMIT25, Usually multiple acids move in and out Transporters 25 |
0.192 |
-0.022 |
| 12 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.192 |
-0.707 |
| 13 |
246719_at |
AT5G28940
|
unknown |
0.188 |
-0.104 |
| 14 |
252598_at |
AT3G45380
|
unknown |
0.187 |
-0.043 |
| 15 |
265485_at |
AT2G15550
|
unknown |
0.182 |
-0.074 |
| 16 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.182 |
-0.433 |
| 17 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
0.181 |
-0.113 |
| 18 |
257360_at |
AT2G39240
|
[RNA polymerase I specific transcription initiation factor RRN3 protein] |
0.181 |
-0.163 |
| 19 |
255761_at |
AT1G16770
|
unknown |
0.180 |
-0.120 |
| 20 |
267615_at |
AT2G26720
|
[Cupredoxin superfamily protein] |
0.180 |
-0.027 |
| 21 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
0.177 |
-0.069 |
| 22 |
265153_at |
AT1G30950
|
UFO, UNUSUAL FLORAL ORGANS |
0.175 |
0.005 |
| 23 |
245979_at |
AT5G13150
|
ATEXO70C1, exocyst subunit exo70 family protein C1, EXO70C1, exocyst subunit exo70 family protein C1 |
0.175 |
-0.115 |
| 24 |
246222_at |
AT4G36900
|
DEAR4, DREB AND EAR MOTIF PROTEIN 4, RAP2.10, related to AP2 10 |
0.169 |
-0.420 |
| 25 |
255557_at |
AT4G01990
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.241 |
0.462 |
| 26 |
265088_at |
AT1G03750
|
CHR9, CHROMATIN REMODELING 9 |
-0.234 |
0.004 |
| 27 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.218 |
0.643 |
| 28 |
246536_at |
AT5G15920
|
EMB2782, EMBRYO DEFECTIVE 2782, SMC5, structural maintenance of chromosomes 5 |
-0.194 |
0.187 |
| 29 |
254681_at |
AT4G18140
|
SSP4b, SCP1-like small phosphatase 4b |
-0.190 |
-0.315 |
| 30 |
246370_at |
AT1G51920
|
unknown |
-0.189 |
0.179 |
| 31 |
245607_at |
AT4G14330
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.187 |
0.657 |
| 32 |
266030_x_at |
AT2G05870
|
[similar to cytochrome P-450 aromatase-related [Arabidopsis thaliana] (TAIR:AT4G07435.1)] |
-0.177 |
-0.117 |
| 33 |
254610_at |
AT4G18890
|
BEH3, BES1/BZR1 homolog 3 |
-0.170 |
0.278 |
| 34 |
252608_at |
AT3G45090
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.166 |
0.419 |
| 35 |
249395_at |
AT5G40190
|
[RNA ligase/cyclic nucleotide phosphodiesterase family protein] |
-0.166 |
0.010 |
| 36 |
260182_at |
AT1G70750
|
MyoB2, myosin binding protein 2 |
-0.163 |
0.098 |
| 37 |
252127_at |
AT3G50960
|
PLP3a, phosducin-like protein 3 homolog |
-0.160 |
0.610 |
| 38 |
250393_at |
AT5G10900
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.150 |
0.095 |
| 39 |
253592_at |
AT4G30840
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.144 |
0.331 |
| 40 |
261628_at |
AT1G50000
|
[methyltransferases] |
-0.137 |
0.055 |
| 41 |
250431_at |
AT5G10440
|
CYCD4;2, cyclin d4;2 |
-0.137 |
-0.059 |
| 42 |
263444_at |
AT2G28700
|
AGL46, AGAMOUS-like 46 |
-0.135 |
-0.018 |
| 43 |
267568_at |
AT2G30780
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.134 |
0.109 |
| 44 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.132 |
-1.019 |
| 45 |
246561_at |
AT5G15570
|
[Bromodomain transcription factor] |
-0.129 |
0.069 |
| 46 |
245383_at |
AT4G17810
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.120 |
-0.435 |
| 47 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.117 |
-0.319 |