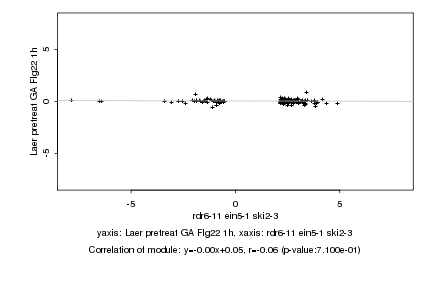
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rdr6-11 ein5-1 ski2-3 |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
AT1G35140 |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
4.865 |
-0.107 |
| 2 |
AT1G50040 |
AT1G50040
|
unknown |
4.317 |
-0.095 |
| 3 |
AT4G30280 |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
4.153 |
0.238 |
| 4 |
AT4G23387 |
AT4G23387
|
MIR845A, microRNA845A |
4.128 |
no data |
| 5 |
AT4G31800 |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
3.911 |
-0.056 |
| 6 |
AT5G52050 |
AT5G52050
|
[MATE efflux family protein] |
3.903 |
-0.030 |
| 7 |
AT1G77640 |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
3.842 |
-0.166 |
| 8 |
AT4G16563 |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
3.806 |
-0.462 |
| 9 |
AT5G57560 |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
3.774 |
0.027 |
| 10 |
AT3G45970 |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
3.770 |
-0.137 |
| 11 |
AT4G25810 |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
3.750 |
0.147 |
| 12 |
AT2G35710 |
AT2G35710
|
PGSIP7, plant glycogenin-like starch initiation protein 7 |
3.626 |
0.055 |
| 13 |
AT1G32170 |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
3.448 |
0.166 |
| 14 |
AT5G01830 |
AT5G01830
|
SAUR21, SMALL AUXIN UP RNA 21 |
3.448 |
0.109 |
| 15 |
AT3G01420 |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
3.389 |
0.885 |
| 16 |
AT1G64340 |
AT1G64340
|
unknown |
3.389 |
no data |
| 17 |
AT1G21910 |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
3.351 |
-0.224 |
| 18 |
AT4G08950 |
AT4G08950
|
EXO, EXORDIUM |
3.342 |
-0.100 |
| 19 |
AT4G28180 |
AT4G28180
|
unknown |
3.336 |
0.024 |
| 20 |
AT4G02330 |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
3.319 |
0.130 |
| 21 |
AT5G41080 |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
3.309 |
0.032 |
| 22 |
AT2G44500 |
AT2G44500
|
[O-fucosyltransferase family protein] |
3.296 |
0.079 |
| 23 |
AT1G73870 |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
3.272 |
-0.292 |
| 24 |
AT5G24030 |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
3.250 |
-0.177 |
| 25 |
AT3G61630 |
AT3G61630
|
CRF6, cytokinin response factor 6 |
3.239 |
-0.062 |
| 26 |
AT2G22850 |
AT2G22850
|
AtbZIP6, basic leucine-zipper 6, bZIP6, basic leucine-zipper 6 |
3.200 |
-0.012 |
| 27 |
AT2G27080 |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
3.191 |
0.125 |
| 28 |
AT3G02140 |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
3.161 |
0.094 |
| 29 |
AT3G60220 |
AT3G60220
|
ATL4, TOXICOS EN LEVADURA 4, TL4, TOXICOS EN LEVADURA 4 |
3.155 |
0.004 |
| 30 |
AT3G51930 |
AT3G51930
|
[Transducin/WD40 repeat-like superfamily protein] |
3.121 |
no data |
| 31 |
AT2G18560 |
AT2G18560
|
[UDP-Glycosyltransferase superfamily protein] |
3.113 |
no data |
| 32 |
AT2G46630 |
AT2G46630
|
unknown |
3.090 |
0.055 |
| 33 |
AT4G37730 |
AT4G37730
|
AtbZIP7, basic leucine-zipper 7, bZIP7, basic leucine-zipper 7 |
3.080 |
0.000 |
| 34 |
AT1G11380 |
AT1G11380
|
[PLAC8 family protein] |
3.018 |
-0.126 |
| 35 |
AT1G56540 |
AT1G56540
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
3.004 |
0.048 |
| 36 |
AT4G19230 |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
2.993 |
0.258 |
| 37 |
AT1G16130 |
AT1G16130
|
WAKL2, wall associated kinase-like 2 |
2.965 |
0.312 |
| 38 |
AT4G08040 |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
2.948 |
0.035 |
| 39 |
AT5G46970 |
AT5G46970
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
2.948 |
no data |
| 40 |
AT1G33760 |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
2.897 |
0.004 |
| 41 |
AT2G41200 |
AT2G41200
|
unknown |
2.896 |
0.019 |
| 42 |
AT3G19680 |
AT3G19680
|
unknown |
2.894 |
-0.088 |
| 43 |
AT3G30775 |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
2.888 |
0.012 |
| 44 |
AT4G18010 |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
2.885 |
-0.035 |
| 45 |
AT3G16860 |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
2.834 |
0.187 |
| 46 |
AT4G04745 |
AT4G04745
|
unknown |
2.803 |
no data |
| 47 |
AT4G33100 |
AT4G33100
|
unknown |
2.802 |
0.171 |
| 48 |
AT5G66070 |
AT5G66070
|
[RING/U-box superfamily protein] |
2.788 |
0.169 |
| 49 |
AT3G02020 |
AT3G02020
|
AK3, aspartate kinase 3 |
2.775 |
no data |
| 50 |
AT2G24600 |
AT2G24600
|
[Ankyrin repeat family protein] |
2.762 |
0.177 |
| 51 |
AT5G11090 |
AT5G11090
|
[serine-rich protein-related] |
2.753 |
0.096 |
| 52 |
AT1G66160 |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
2.753 |
0.033 |
| 53 |
AT3G26605 |
AT3G26605
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 2.4e-51 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
2.751 |
no data |
| 54 |
AT3G19380 |
AT3G19380
|
PUB25, plant U-box 25 |
2.750 |
-0.113 |
| 55 |
AT5G54710 |
AT5G54710
|
[Ankyrin repeat family protein] |
2.741 |
no data |
| 56 |
AT1G13360 |
AT1G13360
|
unknown |
2.715 |
no data |
| 57 |
AT3G57830 |
AT3G57830
|
[Leucine-rich repeat protein kinase family protein] |
2.709 |
0.008 |
| 58 |
AT4G30110 |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
2.691 |
-0.002 |
| 59 |
AT3G10930 |
AT3G10930
|
unknown |
2.686 |
0.087 |
| 60 |
AT4G39190 |
AT4G39190
|
unknown |
2.682 |
0.186 |
| 61 |
AT5G03870 |
AT5G03870
|
[Glutaredoxin family protein] |
2.669 |
0.060 |
| 62 |
AT5G22390 |
AT5G22390
|
unknown |
2.669 |
-0.304 |
| 63 |
AT5G20250 |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
2.662 |
0.072 |
| 64 |
AT5G57220 |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
2.660 |
0.219 |
| 65 |
AT3G43260 |
AT3G43260
|
unknown |
2.655 |
no data |
| 66 |
AT1G22190 |
AT1G22190
|
RAP2.4, related to AP2 4 |
2.652 |
-0.039 |
| 67 |
AT4G24805 |
AT4G24805
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
2.646 |
no data |
| 68 |
AT2G30040 |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
2.619 |
0.118 |
| 69 |
AT5G41750 |
AT5G41750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
2.618 |
no data |
| 70 |
AT4G38400 |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
2.586 |
0.023 |
| 71 |
AT1G55920 |
AT1G55920
|
ATSERAT2;1, serine acetyltransferase 2;1, SAT1, SERINE ACETYLTRANSFERASE 1, SAT5, SERINE ACETYLTRANSFERASE 5, SERAT2;1, serine acetyltransferase 2;1 |
2.579 |
0.087 |
| 72 |
AT3G18700 |
AT3G18700
|
unknown |
2.570 |
0.055 |
| 73 |
AT1G22880 |
AT1G22880
|
ATCEL5, ARABIDOPSIS THALIANA CELLULASE 5, ATGH9B4, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B4, CEL5, cellulase 5 |
2.561 |
0.039 |
| 74 |
AT4G32350 |
AT4G32350
|
[Regulator of Vps4 activity in the MVB pathway protein] |
2.558 |
-0.031 |
| 75 |
ATCG01210 |
ATCG01210
|
RRN16S.2, ribosomal rna 16s |
2.554 |
no data |
| 76 |
AT1G57990 |
AT1G57990
|
ATPUP18, purine permease 18, PUP18, purine permease 18 |
2.546 |
no data |
| 77 |
AT1G19380 |
AT1G19380
|
unknown |
2.546 |
-0.027 |
| 78 |
AT1G69170 |
AT1G69170
|
AtSPL6, SPL6, SQUAMOSA PROMOTER BINDING PROTEIN (SBP)-domain transcription factor 6 |
2.524 |
0.071 |
| 79 |
AT5G49360 |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
2.524 |
-0.162 |
| 80 |
AT4G11000 |
AT4G11000
|
[Ankyrin repeat family protein] |
2.524 |
0.011 |
| 81 |
AT1G54905 |
AT1G54905
|
[copia-like retrotransposon family, has a 4.3e-08 P-value blast match to GB:AAC02666 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
2.523 |
no data |
| 82 |
AT1G02390 |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
2.522 |
0.360 |
| 83 |
AT4G34760 |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
2.496 |
0.099 |
| 84 |
AT3G63110 |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
2.484 |
-0.311 |
| 85 |
AT4G01250 |
AT4G01250
|
AtWRKY22, WRKY22 |
2.477 |
0.061 |
| 86 |
AT3G01290 |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
2.472 |
0.059 |
| 87 |
AT3G55980 |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
2.470 |
-0.126 |
| 88 |
AT2G18570 |
AT2G18570
|
[UDP-Glycosyltransferase superfamily protein] |
2.463 |
no data |
| 89 |
AT5G48850 |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
2.459 |
0.110 |
| 90 |
AT3G47500 |
AT3G47500
|
CDF3, cycling DOF factor 3 |
2.453 |
0.026 |
| 91 |
AT4G14370 |
AT4G14370
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
2.444 |
no data |
| 92 |
AT5G01100 |
AT5G01100
|
FRB1, FRIABLE 1 |
2.444 |
no data |
| 93 |
AT1G16110 |
AT1G16110
|
WAKL6, wall associated kinase-like 6 |
2.436 |
no data |
| 94 |
AT1G61667 |
AT1G61667
|
unknown |
2.435 |
0.011 |
| 95 |
AT2G01080 |
AT2G01080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
2.425 |
-0.077 |
| 96 |
AT1G67635 |
AT1G67635
|
unknown |
2.409 |
0.073 |
| 97 |
AT4G00970 |
AT4G00970
|
CRK41, cysteine-rich RLK (RECEPTOR-like protein kinase) 41 |
2.397 |
0.166 |
| 98 |
AT5G45340 |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
2.396 |
0.264 |
| 99 |
AT3G44935 |
AT3G44935
|
unknown |
2.393 |
no data |
| 100 |
AT1G76650 |
AT1G76650
|
CML38, calmodulin-like 38 |
2.383 |
0.198 |
| 101 |
AT2G23290 |
AT2G23290
|
AtMYB70, myb domain protein 70, MYB70, myb domain protein 70 |
2.381 |
-0.063 |
| 102 |
AT4G32800 |
AT4G32800
|
[Integrase-type DNA-binding superfamily protein] |
2.376 |
-0.069 |
| 103 |
AT5G49170 |
AT5G49170
|
unknown |
2.374 |
0.115 |
| 104 |
AT3G29670 |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
2.374 |
0.041 |
| 105 |
AT3G16510 |
AT3G16510
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
2.356 |
no data |
| 106 |
AT1G78080 |
AT1G78080
|
AtWIND1, RAP2.4, related to AP2 4, WIND1, wound induced dedifferentiation 1 |
2.349 |
0.046 |
| 107 |
AT1G12020 |
AT1G12020
|
unknown |
2.344 |
-0.114 |
| 108 |
AT3G55840 |
AT3G55840
|
[Hs1pro-1 protein] |
2.334 |
0.238 |
| 109 |
AT4G13340 |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
2.323 |
0.059 |
| 110 |
AT5G64870 |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
2.320 |
0.322 |
| 111 |
AT1G19020 |
AT1G19020
|
unknown |
2.317 |
0.179 |
| 112 |
AT1G20070 |
AT1G20070
|
unknown |
2.316 |
-0.112 |
| 113 |
AT2G38470 |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
2.311 |
0.052 |
| 114 |
AT3G49580 |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
2.311 |
0.276 |
| 115 |
AT4G23810 |
AT4G23810
|
ATWRKY53, WRKY53 |
2.302 |
-0.038 |
| 116 |
AT2G35930 |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
2.290 |
0.069 |
| 117 |
AT4G17460 |
AT4G17460
|
HAT1, JAB, JAIBA |
2.284 |
-0.272 |
| 118 |
AT3G50890 |
AT3G50890
|
AtHB28, homeobox protein 28, HB28, homeobox protein 28, ZHD7, ZINC FINGER HOMEODOMAIN 7 |
2.281 |
0.066 |
| 119 |
AT4G33985 |
AT4G33985
|
unknown |
2.278 |
0.270 |
| 120 |
AT3G42800 |
AT3G42800
|
unknown |
2.271 |
0.046 |
| 121 |
AT1G03610 |
AT1G03610
|
unknown |
2.264 |
0.048 |
| 122 |
AT3G59080 |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
2.237 |
0.189 |
| 123 |
AT1G10340 |
AT1G10340
|
[Ankyrin repeat family protein] |
2.236 |
0.047 |
| 124 |
AT1G19220 |
AT1G19220
|
ARF11, AUXIN RESPONSE FACTOR11, ARF19, auxin response factor 19, IAA22, indole-3-acetic acid inducible 22 |
2.236 |
-0.044 |
| 125 |
AT5G36925 |
AT5G36925
|
unknown |
2.236 |
no data |
| 126 |
AT2G36470 |
AT2G36470
|
unknown |
2.225 |
0.006 |
| 127 |
AT1G07135 |
AT1G07135
|
[glycine-rich protein] |
2.224 |
no data |
| 128 |
AT3G59350 |
AT3G59350
|
[Protein kinase superfamily protein] |
2.223 |
0.376 |
| 129 |
AT2G17230 |
AT2G17230
|
EXL5, EXORDIUM like 5 |
2.217 |
-0.050 |
| 130 |
AT5G56980 |
AT5G56980
|
unknown |
2.212 |
0.166 |
| 131 |
AT1G72790 |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
2.210 |
0.133 |
| 132 |
AT3G08720 |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
2.202 |
0.123 |
| 133 |
AT3G63450 |
AT3G63450
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
2.196 |
no data |
| 134 |
AT3G24270 |
AT3G24270
|
APUM25, pumilio 25, PUM25, pumilio 25 |
2.190 |
0.069 |
| 135 |
AT5G66650 |
AT5G66650
|
unknown |
2.176 |
0.168 |
| 136 |
AT5G52750 |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
2.174 |
0.111 |
| 137 |
AT3G05490 |
AT3G05490
|
RALFL22, ralf-like 22 |
2.170 |
0.010 |
| 138 |
AT2G41100 |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
2.156 |
0.089 |
| 139 |
AT4G04632 |
AT4G04632
|
[Protein kinase superfamily protein] |
2.155 |
no data |
| 140 |
AT4G37250 |
AT4G37250
|
[Leucine-rich repeat protein kinase family protein] |
2.152 |
-0.005 |
| 141 |
AT1G28370 |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
2.151 |
-0.119 |
| 142 |
AT3G06070 |
AT3G06070
|
unknown |
2.149 |
-0.141 |
| 143 |
AT1G63860 |
AT1G63860
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
2.147 |
no data |
| 144 |
AT3G50800 |
AT3G50800
|
unknown |
2.139 |
no data |
| 145 |
AT3G62690 |
AT3G62690
|
ATL5, AtL5 |
2.127 |
0.014 |
| 146 |
AT2G23690 |
AT2G23690
|
unknown |
2.126 |
no data |
| 147 |
AT1G62045 |
AT1G62045
|
unknown |
2.126 |
-0.068 |
| 148 |
AT2G36090 |
AT2G36090
|
[F-box family protein] |
2.126 |
0.225 |
| 149 |
AT2G01300 |
AT2G01300
|
unknown |
2.126 |
0.389 |
| 150 |
AT3G44580 |
AT3G44580
|
unknown |
-7.889 |
0.103 |
| 151 |
AT3G44570 |
AT3G44570
|
[Arabidopsis retrotransposon ORF-1 protein] |
-6.549 |
0.028 |
| 152 |
AT5G42980 |
AT5G42980
|
ATH3, thioredoxin H-type 3, ATTRX3, thioredoxin 3, ATTRXH3, TRX3, thioredoxin 3, TRXH3, THIOREDOXIN H3 |
-6.449 |
0.039 |
| 153 |
ATCG01160 |
ATCG01160
|
RRN5S, ribosomal RNA5S |
-6.230 |
no data |
| 154 |
AT1G50055 |
AT1G50055
|
TAS1B, trans-acting siRNA1B |
-5.640 |
no data |
| 155 |
AT3G25795 |
AT3G25795
|
TAS4, trans acting siRNA 4 |
-5.058 |
no data |
| 156 |
ATCG00960 |
ATCG00960
|
RRN4.5S.1, ribosomal RNA4.5S |
-3.572 |
no data |
| 157 |
AT1G63630 |
AT1G63630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-3.485 |
no data |
| 158 |
AT1G63150 |
AT1G63150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-3.437 |
0.032 |
| 159 |
AT3G49500 |
AT3G49500
|
RDR6, RNA-dependent RNA polymerase 6, SDE1, SILENCING DEFECTIVE 1, SGS2, SUPPRESSOR OF GENE SILENCING 2 |
-3.082 |
-0.037 |
| 160 |
AT5G44430 |
AT5G44430
|
PDF1.2c, plant defensin 1.2C |
-2.960 |
no data |
| 161 |
AT2G26010 |
AT2G26010
|
PDF1.3, plant defensin 1.3 |
-2.914 |
no data |
| 162 |
AT1G63130 |
AT1G63130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-2.766 |
0.054 |
| 163 |
AT3G49620 |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-2.766 |
0.078 |
| 164 |
AT5G49615 |
AT5G49615
|
TAS3b, trans-acting siRNA3B |
-2.749 |
no data |
| 165 |
AT1G63080 |
AT1G63080
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-2.567 |
0.097 |
| 166 |
AT1G62910 |
AT1G62910
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-2.547 |
no data |
| 167 |
AT5G57735 |
AT5G57735
|
TASIR-ARF |
-2.529 |
no data |
| 168 |
AT5G44420 |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-2.428 |
-0.171 |
| 169 |
AT1G53480 |
AT1G53480
|
ATMRD1, ARABIDOPSIS MTO 1 RESPONDING DOWN 1, MRD1, MTO 1 RESPONDING DOWN 1 |
-2.319 |
no data |
| 170 |
AT1G24068 |
AT1G24068
|
other RNA |
-2.169 |
no data |
| 171 |
AT2G40680 |
AT2G40680
|
unknown |
-2.087 |
0.129 |
| 172 |
AT4G08100 |
AT4G08100
|
[gypsy-like retrotransposon family, has a 1.2e-123 P-value blast match to GB:AAD22153 polyprotein (Gypsy_Ty3-element) (Sorghum bicolor)] |
-2.060 |
no data |
| 173 |
AT3G52705 |
AT3G52705
|
[copia-like retrotransposon family, has a 7.3e-24 P-value blast match to reverse transcriptase (Ty1_Copia-element) (Arabidopsis thaliana)] |
-1.996 |
no data |
| 174 |
AT1G62930 |
AT1G62930
|
RPF3, RNA processing factor 3 |
-1.987 |
0.039 |
| 175 |
AT2G30770 |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
-1.933 |
0.691 |
| 176 |
AT5G38850 |
AT5G38850
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-1.910 |
0.115 |
| 177 |
AT2G39030 |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-1.875 |
0.108 |
| 178 |
AT1G61120 |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
-1.875 |
0.026 |
| 179 |
AT3G17185 |
AT3G17185
|
ATTAS3, TAS3, trans-acting siRNA3, TASIR-ARF |
-1.825 |
no data |
| 180 |
AT5G36228 |
AT5G36228
|
[nucleic acid binding] |
-1.768 |
no data |
| 181 |
AT5G35935 |
AT5G35935
|
[copia-like retrotransposon family, has a 2.0e-232 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-1.768 |
no data |
| 182 |
AT3G49070 |
AT3G49070
|
unknown |
-1.765 |
0.103 |
| 183 |
AT3G07380 |
AT3G07380
|
unknown |
-1.731 |
no data |
| 184 |
AT4G36925 |
AT4G36925
|
unknown |
-1.731 |
no data |
| 185 |
AT2G29880 |
AT2G29880
|
unknown |
-1.715 |
0.065 |
| 186 |
AT1G06148 |
AT1G06148
|
unknown |
-1.699 |
no data |
| 187 |
AT5G61160 |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-1.614 |
-0.019 |
| 188 |
AT3G50540 |
AT3G50540
|
unknown |
-1.584 |
no data |
| 189 |
AT4G29090 |
AT4G29090
|
[Ribonuclease H-like superfamily protein] |
-1.567 |
0.089 |
| 190 |
AT3G16530 |
AT3G16530
|
[Legume lectin family protein] |
-1.513 |
0.185 |
| 191 |
AT3G54925 |
AT3G54925
|
[Plant self-incompatibility protein S1 family] |
-1.478 |
no data |
| 192 |
AT1G33300 |
AT1G33300
|
[pseudogene] |
-1.474 |
no data |
| 193 |
AT3G24260 |
AT3G24260
|
unknown |
-1.444 |
0.003 |
| 194 |
AT4G16260 |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
-1.432 |
0.244 |
| 195 |
AT5G20780 |
AT5G20780
|
[Mutator-like transposase family, has a 3.4e-66 P-value blast match to Q9ZQM3 /24-192 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-1.425 |
no data |
| 196 |
AT3G25460 |
AT3G25460
|
[F-box and associated interaction domains-containing protein] |
-1.425 |
no data |
| 197 |
AT1G69530 |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-1.382 |
0.291 |
| 198 |
AT3G23550 |
AT3G23550
|
[MATE efflux family protein] |
-1.356 |
-0.035 |
| 199 |
AT2G16560 |
AT2G16560
|
[non-LTR retrotransposon family (LINE), has a 1.7e-41 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-1.300 |
no data |
| 200 |
AT2G13490 |
AT2G13490
|
[non-LTR retrotransposon family (LINE), has a 3.3e-19 P-value blast match to GB:NP_038607 L1 repeat, Tf subfamily, member 9 (LINE-element) (Mus musculus)] |
-1.300 |
no data |
| 201 |
AT4G06552 |
AT4G06552
|
[Mutator-like transposase family, has a 8.6e-69 P-value blast match to O80466 /172-336 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-1.298 |
no data |
| 202 |
AT2G45245 |
AT2G45245
|
other RNA |
-1.276 |
no data |
| 203 |
AT3G48526 |
AT3G48526
|
[copia-like retrotransposon family, has a 5.7e-41 P-value blast match to GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)gi|4996361|dbj|BAA78423.1| polyprotein (Arabidopsis thaliana) (Ty1_Copia-element)] |
-1.273 |
no data |
| 204 |
AT3G12500 |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
-1.214 |
0.289 |
| 205 |
AT3G15356 |
AT3G15356
|
[Legume lectin family protein] |
-1.211 |
no data |
| 206 |
AT3G04720 |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
-1.159 |
0.160 |
| 207 |
AT1G19670 |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-1.140 |
-0.483 |
| 208 |
AT5G15833 |
AT5G15833
|
MIR865A, microRNA865A |
-1.126 |
no data |
| 209 |
AT2G13450 |
AT2G13450
|
unknown |
-1.084 |
no data |
| 210 |
AT4G10990 |
AT4G10990
|
[copia-like retrotransposon family, has a 1.2e-190 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-1.083 |
0.046 |
| 211 |
AT2G17115 |
AT2G17115
|
[copia-like retrotransposon family, has a 1.3e-251 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-1.079 |
no data |
| 212 |
AT1G09486 |
AT1G09486
|
[pseudogene] |
-1.071 |
no data |
| 213 |
AT1G08105 |
AT1G08105
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 1.0e-298 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
-1.069 |
no data |
| 214 |
AT1G48953 |
AT1G48953
|
unknown |
-1.069 |
no data |
| 215 |
AT5G10945 |
AT5G10945
|
MIR156D, microRNA156D |
-1.062 |
no data |
| 216 |
AT4G34071 |
AT4G34071
|
other RNA |
-1.053 |
no data |
| 217 |
AT4G22415 |
AT4G22415
|
[gypsy-like retrotransposon family, has a 1.8e-181 P-value blast match to GB:AAD22153 polyprotein (Gypsy_Ty3-element) (Sorghum bicolor)] |
-1.049 |
no data |
| 218 |
AT1G65585 |
AT1G65585
|
[pseudogene] |
-1.040 |
no data |
| 219 |
AT1G70050 |
AT1G70050
|
[tRNA-Pro (anticodon: AGG)] |
-1.038 |
no data |
| 220 |
AT3G26483 |
AT3G26483
|
[copia-like retrotransposon family, has a 3.4e-252 P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-1.029 |
no data |
| 221 |
AT3G16670 |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-1.011 |
0.106 |
| 222 |
AT2G19100 |
AT2G19100
|
[non-LTR retrotransposon family (LINE), has a 3.2e-33 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
-1.008 |
0.054 |
| 223 |
AT2G06240 |
AT2G06240
|
[Mutator-like transposase family, has a 3.7e-37 P-value blast match to Q9SI25 /181-349 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.982 |
no data |
| 224 |
AT3G43686 |
AT3G43686
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 2.0e-142 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
-0.973 |
no data |
| 225 |
AT2G16676 |
AT2G16676
|
unknown |
-0.969 |
no data |
| 226 |
AT1G54490 |
AT1G54490
|
AIN1, ACC INSENSITIVE 1, ATXRN4, EXORIBONUCLEASE 4, EIN5, ETHYLENE INSENSITIVE 5, XRN4, exoribonuclease 4 |
-0.962 |
0.044 |
| 227 |
AT3G43780 |
AT3G43780
|
[pseudogene] |
-0.961 |
no data |
| 228 |
AT2G18042 |
AT2G18042
|
|
-0.948 |
no data |
| 229 |
AT2G41180 |
AT2G41180
|
SIB2, sigma factor binding protein 2 |
-0.947 |
-0.309 |
| 230 |
AT4G06548 |
AT4G06548
|
[Mutator-like transposase family, has a 2.5e-39 P-value blast match to Q9SJR8 /172-333 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.942 |
no data |
| 231 |
AT3G44990 |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.941 |
-0.024 |
| 232 |
AT4G27300 |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.941 |
-0.329 |
| 233 |
AT2G33920 |
AT2G33920
|
[tRNA-Pro (anticodon: AGG)] |
-0.940 |
no data |
| 234 |
AT2G03700 |
AT2G03700
|
[tRNA-Thr (anticodon: CGT)] |
-0.937 |
no data |
| 235 |
AT5G27882 |
AT5G27882
|
[copia-like retrotransposon family, has a 1.5e-36 P-value blast match to GB:AAC02666 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
-0.921 |
no data |
| 236 |
AT5G45605 |
AT5G45605
|
[copia-like retrotransposon family, has a 9.0e-230 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.906 |
no data |
| 237 |
AT2G15410 |
AT2G15410
|
[gypsy-like retrotransposon family, has a 4.1e-217 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.903 |
0.117 |
| 238 |
AT2G30180 |
AT2G30180
|
[tRNA-Gly (anticodon: GCC)] |
-0.898 |
no data |
| 239 |
ATCG00970 |
ATCG00970
|
RRN5S, ribosomal RNA5S |
-0.848 |
no data |
| 240 |
AT3G29515 |
AT3G29515
|
[non-LTR retrotransposon family (LINE), has a 1.6e-11 P-value blast match to GB:NP_038602 L1 repeat, Tf subfamily, member 18 (LINE-element) (Mus musculus)] |
-0.847 |
no data |
| 241 |
AT5G43740 |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.844 |
0.073 |
| 242 |
AT5G53135 |
AT5G53135
|
[copia-like retrotransposon family, has a 6.9e-199 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.839 |
no data |
| 243 |
AT1G29650 |
AT1G29650
|
[non-LTR retrotransposon family (LINE), has a 1.5e-28 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-0.833 |
0.093 |
| 244 |
AT1G07051 |
AT1G07051
|
MIR847A, microRNA847A |
-0.830 |
no data |
| 245 |
AT2G19840 |
AT2G19840
|
[copia-like retrotransposon family, has a 3.5e-301 P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
-0.826 |
0.028 |
| 246 |
AT3G10745 |
AT3G10745
|
MIR158A, microRNA158A |
-0.823 |
no data |
| 247 |
AT3G16770 |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.799 |
-0.115 |
| 248 |
AT2G16680 |
AT2G16680
|
[non-LTR retrotransposon family (LINE), has a 5.0e-35 P-value blast match to GB:NP_038605 L1 repeat, Tf subfamily, member 30 (LINE-element) (Mus musculus)] |
-0.793 |
0.060 |
| 249 |
AT4G04560 |
AT4G04560
|
[copia-like retrotransposon family, has a 1.0e-94 P-value blast match to gb|AAG52949.1| gag/pol polyprotein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
-0.793 |
0.047 |
| 250 |
AT3G50625 |
AT3G50625
|
[copia-like retrotransposon family, has a 6.1e-96 P-value blast match to dbj|BAA78425.1| polyprotein (Arabidopsis thaliana) (AtRE1) (Ty1_Copia-element)] |
-0.790 |
no data |
| 251 |
AT3G43828 |
AT3G43828
|
[CACTA-like transposase family (En/Spm), has a 5.8e-16 P-value blast match to GB:BAA20532 ORF of transposon Tdc1 (CACTA-element) (Daucus carota)] |
-0.785 |
no data |
| 252 |
AT1G67240 |
AT1G67240
|
[Mutator-like transposase family, has a 4.5e-23 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.783 |
-0.009 |
| 253 |
AT2G05935 |
AT2G05935
|
[non-LTR retrotransposon family (LINE), has a 3.9e-28 P-value blast match to GB:NP_038607 L1 repeat, Tf subfamily, member 9 (LINE-element) (Mus musculus)] |
-0.780 |
no data |
| 254 |
AT1G31163 |
AT1G31163
|
[F-box associated ubiquitination effector family protein] |
-0.778 |
no data |
| 255 |
AT1G70010 |
AT1G70010
|
[copia-like retrotransposon family, has a 3.2e-289 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.769 |
no data |
| 256 |
AT2G04400 |
AT2G04400
|
[Aldolase-type TIM barrel family protein] |
-0.768 |
0.185 |
| 257 |
AT5G64770 |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.764 |
0.185 |
| 258 |
AT2G21460 |
AT2G21460
|
[copia-like retrotransposon family, has a 1.3e-303 P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
-0.762 |
0.079 |
| 259 |
AT1G54430 |
AT1G54430
|
unknown |
-0.758 |
no data |
| 260 |
AT2G45790 |
AT2G45790
|
ATPMM, PHOSPHOMANNOMUTASE, PMM, phosphomannomutase |
-0.755 |
0.141 |
| 261 |
AT4G34630 |
AT4G34630
|
unknown |
-0.751 |
0.093 |
| 262 |
AT1G48422 |
AT1G48422
|
|
-0.741 |
no data |
| 263 |
AT2G46255 |
AT2G46255
|
MIR159C, microRNA159C |
-0.721 |
no data |
| 264 |
AT1G01270 |
AT1G01270
|
[tRNA-Gln (anticodon: CTG)] |
-0.721 |
no data |
| 265 |
AT1G24967 |
AT1G24967
|
[Mutator-like transposase family, has a 1.2e-61 P-value blast match to Q9S9L6 /322-461 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.720 |
no data |
| 266 |
AT2G34900 |
AT2G34900
|
GTE01, GTE1, GLOBAL TRANSCRIPTION FACTOR GROUP E1, IMB1, IMBIBITION-INDUCIBLE 1 |
-0.712 |
-0.092 |
| 267 |
AT3G17400 |
AT3G17400
|
[F-box family protein] |
-0.707 |
no data |
| 268 |
AT2G43138 |
AT2G43138
|
[snoRNA] |
-0.704 |
no data |
| 269 |
AT5G29720 |
AT5G29720
|
[gypsy-like retrotransposon family (Athila), has a 2.7e-94 P-value blast match to gb|AAL06421.1|AF378079_1 reverse transcriptase (Athila4) (Arabidopsis thaliana) (Gypsy_Ty3-family)] |
-0.693 |
no data |
| 270 |
AT1G48267 |
AT1G48267
|
MIR161, microRNA161 |
-0.692 |
no data |
| 271 |
AT5G26775 |
AT5G26775
|
[non-LTR retrotransposon family (LINE), has a 5.9e-28 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
-0.690 |
no data |
| 272 |
AT3G26775 |
AT3G26775
|
[pseudogene] |
-0.689 |
no data |
| 273 |
AT3G21870 |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.684 |
-0.086 |
| 274 |
AT4G00120 |
AT4G00120
|
EDA33, EMBRYO SAC DEVELOPMENT ARREST 33, GT140, IND, INDEHISCENT, IND1, INDEHISCENT |
-0.651 |
no data |
| 275 |
AT1G61226 |
AT1G61226
|
MIR846A, microRNA846A |
-0.650 |
no data |
| 276 |
AT5G31770 |
AT5G31770
|
[gypsy-like retrotransposon family (Athila), has a 3.7e-53 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.647 |
no data |
| 277 |
AT1G18075 |
AT1G18075
|
MIR159, MICRORNA 159, MIR159B, microRNA159B |
-0.632 |
no data |
| 278 |
AT5G47815 |
AT5G47815
|
[non-LTR retrotransposon family (LINE), has a 3.0e-38 P-value blast match to GB:NP_038602 L1 repeat, Tf subfamily, member 18 (LINE-element) (Mus musculus)] |
-0.627 |
no data |
| 279 |
AT3G42240 |
AT3G42240
|
unknown |
-0.627 |
0.036 |
| 280 |
AT1G58561 |
AT1G58561
|
[copia-like retrotransposon family, has a 3.8e-219 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
-0.620 |
no data |
| 281 |
AT1G55591 |
AT1G55591
|
MIR158B, microRNA158B |
-0.616 |
no data |
| 282 |
AT5G40395 |
AT5G40395
|
U6acat |
-0.616 |
no data |
| 283 |
AT3G28925 |
AT3G28925
|
unknown |
-0.611 |
no data |
| 284 |
AT2G16670 |
AT2G16670
|
[copia-like retrotransposon family, has a 1.2e-190 P-value blast match to GB:AAB82754 retrofit (TY1_Copia-element) (Oryza longistaminata)] |
-0.606 |
-0.038 |
| 285 |
AT2G31080 |
AT2G31080
|
[non-LTR retrotransposon family (LINE), has a 1.3e-49 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-0.604 |
0.034 |
| 286 |
AT1G73687 |
AT1G73687
|
MIR159, MICRORNA 159, MIR159A, microRNA159A |
-0.602 |
no data |
| 287 |
AT2G43141 |
AT2G43141
|
[snoRNA] |
-0.591 |
no data |
| 288 |
AT2G44570 |
AT2G44570
|
AtGH9B12, glycosyl hydrolase 9B12, GH9B12, glycosyl hydrolase 9B12 |
-0.587 |
0.019 |
| 289 |
AT5G35643 |
AT5G35643
|
[pseudogene] |
-0.586 |
no data |
| 290 |
AT4G01490 |
AT4G01490
|
[non-LTR retrotransposon family (LINE), has a 2.4e-44 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
-0.571 |
0.046 |
| 291 |
AT1G17570 |
AT1G17570
|
[tRNA-Ser (anticodon: AGA)] |
-0.564 |
no data |
| 292 |
AT2G33950 |
AT2G33950
|
[tRNA-Pro (anticodon: CGG)] |
-0.563 |
no data |
| 293 |
AT5G54960 |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
-0.555 |
0.089 |
| 294 |
AT3G42433 |
AT3G42433
|
[copia-like retrotransposon family, has a 0.00033 P-value blast match to GB:AAD12998 pol polyprotein (Ty1_Copia-element) (Zea mays)] |
-0.553 |
no data |
| 295 |
AT2G43150 |
AT2G43150
|
[Proline-rich extensin-like family protein] |
-0.553 |
0.026 |
| 296 |
AT3G22555 |
AT3G22555
|
[pseudogene] |
-0.545 |
no data |
| 297 |
AT5G38365 |
AT5G38365
|
[non-LTR retrotransposon family (LINE), has a 6.0e-32 P-value blast match to GB:NP_038605 L1 repeat, Tf subfamily, member 30 (LINE-element) (Mus musculus)] |
-0.535 |
no data |